The structure of the TH/INS locus and the parental allele expressed are not conserved between mammals
- PMID: 38834866
- PMCID: PMC11222543
- DOI: 10.1038/s41437-024-00689-y
The structure of the TH/INS locus and the parental allele expressed are not conserved between mammals
Abstract
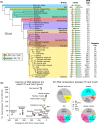
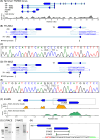
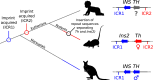
Parent-of-origin-specific expression of imprinted genes is critical for successful mammalian growth and development. Insulin, coded by the INS gene, is an important growth factor expressed from the paternal allele in the yolk sac placenta of therian mammals. The tyrosine hydroxylase gene TH encodes an enzyme involved in dopamine synthesis. TH and INS are closely associated in most vertebrates, but the mouse orthologues, Th and Ins2, are separated by repeated DNA. In mice, Th is expressed from the maternal allele, but the parental origin of expression is not known for any other mammal so it is unclear whether the maternal expression observed in the mouse represents an evolutionary divergence or an ancestral condition. We compared the length of the DNA segment between TH and INS across species and show that separation of these genes occurred in the rodent lineage with an accumulation of repeated DNA. We found that the region containing TH and INS in the tammar wallaby produces at least five distinct RNA transcripts: TH, TH-INS1, TH-INS2, lncINS and INS. Using allele-specific expression analysis, we show that the TH/INS locus is expressed from the paternal allele in pre- and postnatal tammar wallaby tissues. Determining the imprinting pattern of TH/INS in other mammals might clarify if paternal expression is the ancestral condition which has been flipped to maternal expression in rodents by the accumulation of repeat sequences.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Evidence that insulin is imprinted in the human yolk sac.Diabetes. 2001 Jan;50(1):199-203. doi: 10.2337/diabetes.50.1.199. Diabetes. 2001. PMID: 11147788
-
Characterisation of marsupial PHLDA2 reveals eutherian specific acquisition of imprinting.BMC Evol Biol. 2011 Aug 19;11:244. doi: 10.1186/1471-2148-11-244. BMC Evol Biol. 2011. PMID: 21854573 Free PMC article.
-
Differential expression and imprinting status of Ins1 and Ins2 genes in extraembryonic tissues of laboratory mice.Gene Expr Patterns. 2004 Dec;5(2):297-300. doi: 10.1016/j.modgep.2004.04.013. Gene Expr Patterns. 2004. PMID: 15567729
-
The conflict theory of genomic imprinting: how much can be explained?Curr Top Dev Biol. 1998;40:255-93. doi: 10.1016/s0070-2153(08)60369-5. Curr Top Dev Biol. 1998. PMID: 9673853 Review.
-
Genomic imprinting in mice: its function and mechanism.Biol Reprod. 1996 Feb;54(2):273-8. doi: 10.1095/biolreprod54.2.273. Biol Reprod. 1996. PMID: 8788176 Review.
Cited by
-
PRKACB is a novel imprinted gene in marsupials.Epigenetics Chromatin. 2024 Sep 28;17(1):29. doi: 10.1186/s13072-024-00552-8. Epigenetics Chromatin. 2024. PMID: 39342354 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

