Epigenetic and proteomic signatures associate with clonal hematopoiesis expansion rate
- PMID: 38834882
- PMCID: PMC11832052
- DOI: 10.1038/s43587-024-00647-7
Epigenetic and proteomic signatures associate with clonal hematopoiesis expansion rate
Abstract
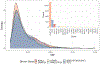
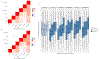
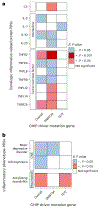
Clonal hematopoiesis of indeterminate potential (CHIP), whereby somatic mutations in hematopoietic stem cells confer a selective advantage and drive clonal expansion, not only correlates with age but also confers increased risk of morbidity and mortality. Here, we leverage genetically predicted traits to identify factors that determine CHIP clonal expansion rate. We used the passenger-approximated clonal expansion rate method to quantify the clonal expansion rate for 4,370 individuals in the National Heart, Lung, and Blood Institute (NHLBI) Trans-Omics for Precision Medicine (TOPMed) cohort and calculated polygenic risk scores for DNA methylation aging, inflammation-related measures and circulating protein levels. Clonal expansion rate was significantly associated with both genetically predicted and measured epigenetic clocks. No associations were identified with inflammation-related lab values or diseases and CHIP expansion rate overall. A proteome-wide search identified predicted circulating levels of myeloid zinc finger 1 and anti-Müllerian hormone as associated with an increased CHIP clonal expansion rate and tissue inhibitor of metalloproteinase 1 and glycine N-methyltransferase as associated with decreased CHIP clonal expansion rate. Together, our findings identify epigenetic and proteomic patterns associated with the rate of hematopoietic clonal expansion.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
S.J. is on advisory boards for Novartis, AVRO Bio, and Roche Genentech, reports speaking fees and a honorarium from GSK, and is on the scientific advisory board of Bitterroot Bio. P.N. reports grants support from Amgen, AstraZeneca, Apple, Novartis and Boston Scientific, is a paid consultant for Apple, AstraZeneca, Novartis, Genentech, Blackstone Life Sciences and spousal employment at Vertex, all unrelated to the present work. S.J., A.G.B. and P.N. are paid consultants for Foresite Labs and co-founders, equity holders, and on the scientific advisory board of TenSixteen Bio. Stanford University has filed a patent application for the use of PACER to identify therapeutic targets on which S.J., A.G.B. and J.S.W. are inventors (US patent 63/141,333). The patent has been licensed to TenSixteen Bio. A.S. is an employee of Regeneron Pharmaceuticals. B.M.P. serves on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. E.S. receives grant support from Bayer and GSK. J.Y. & D.D. receive grant support from Bayer. M.C. receives grant support from Bayer and GSK, and consulting and speaking fees from Illumina and AstraZeneca. S.S.R. & L.M.R. are paid consultants for Westat, the Administrative Coordinating Center for the NHLBI Trans-Omics for Precision Medicine (TOPMed) program. The remaining authors declare no competing interests.
Figures

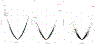


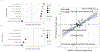

References
-
- Argüelles OC et al. Clonal hematopoiesis associated mutations, cardiovascular events, and all-cause death among patients with acute myeloid leukemia. J. Am. Coll. Cardiol 75, 671–671 (2020).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

