Cell type mapping of inflammatory muscle diseases highlights selective myofiber vulnerability in inclusion body myositis
- PMID: 38834884
- PMCID: PMC11257986
- DOI: 10.1038/s43587-024-00645-9
Cell type mapping of inflammatory muscle diseases highlights selective myofiber vulnerability in inclusion body myositis
Abstract
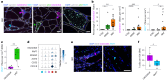
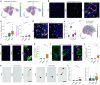
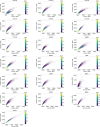
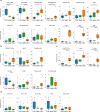
Inclusion body myositis (IBM) is the most prevalent inflammatory muscle disease in older adults with no effective therapy available. In contrast to other inflammatory myopathies such as subacute, immune-mediated necrotizing myopathy (IMNM), IBM follows a chronic disease course with both inflammatory and degenerative features of pathology. Moreover, causal factors and molecular drivers of IBM progression are largely unknown. Therefore, we paired single-nucleus RNA sequencing with spatial transcriptomics from patient muscle biopsies to map cell-type-specific drivers underlying IBM pathogenesis compared with IMNM muscles and noninflammatory skeletal muscle samples. In IBM muscles, we observed a selective loss of type 2 myonuclei paralleled by increased levels of cytotoxic T and conventional type 1 dendritic cells. IBM myofibers were characterized by either upregulation of cell stress markers featuring GADD45A and NORAD or protein degradation markers including RNF7 associated with p62 aggregates. GADD45A upregulation was preferentially seen in type 2A myofibers associated with severe tissue inflammation. We also noted IBM-specific upregulation of ACHE encoding acetylcholinesterase, which can be regulated by NORAD activity and result in functional denervation of myofibers. Our results provide promising insights into possible mechanisms of myofiber degeneration in IBM and suggest a selective type 2 fiber vulnerability linked to genomic stress and denervation pathways.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

