Tumor antigen PRAME is a potential therapeutic target of p53 activation in melanoma cells
- PMID: 38835116
- PMCID: PMC11214892
- DOI: 10.5483/BMBRep.2023-0246
Tumor antigen PRAME is a potential therapeutic target of p53 activation in melanoma cells
Abstract
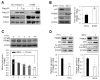
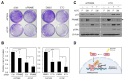
Upregulation of PRAME (preferentially expressed antigen of melanoma) has been implicated in the progression of a variety of cancers, including melanoma. The tumor suppressor p53 is a transcriptional regulator that mediates cell cycle arrest and apoptosis in response to stress signals. Here, we report that PRAME is a novel repressive target of p53. This was supported by analysis of melanoma cell lines carrying wild-type p53 and human melanoma databases. mRNA expression of PRAME was downregulated by p53 overexpression and activation using DNA-damaging agents, but upregulated by p53 depletion. We identified a p53-responsive element (p53RE) in the promoter region of PRAME. Luciferase and ChIP assays showed that p53 represses the transcriptional activity of the PRAME promoter and is recruited to the p53RE together with HDAC1 upon etoposide treatment. The functional significance of p53 activationmediated PRAME downregulation was demonstrated by measuring colony formation and p27 expression in melanoma cells. These data suggest that p53 activation, which leads to PRAME downregulation, could be a therapeutic strategy in melanoma cells. [BMB Reports 2024; 57(6): 299-304].
Conflict of interest statement
The authors have no conflicting interests.
Figures


Similar articles
-
Tumor antigen PRAME is up-regulated by MZF1 in cooperation with DNA hypomethylation in melanoma cells.Cancer Lett. 2017 Sep 10;403:144-151. doi: 10.1016/j.canlet.2017.06.015. Epub 2017 Jun 17. Cancer Lett. 2017. PMID: 28634046
-
Downregulation of microRNA-211 is involved in expression of preferentially expressed antigen of melanoma in melanoma cells.Int J Oncol. 2011 Sep;39(3):665-72. doi: 10.3892/ijo.2011.1084. Epub 2011 Jun 16. Int J Oncol. 2011. PMID: 21687938
-
Downregulation of PRAME Suppresses Proliferation and Promotes Apoptosis in Hepatocellular Carcinoma Through the Activation of P53 Mediated Pathway.Cell Physiol Biochem. 2018;45(3):1121-1135. doi: 10.1159/000487353. Epub 2018 Feb 7. Cell Physiol Biochem. 2018. PMID: 29439259
-
PRAME Updated: Diagnostic, Prognostic, and Therapeutic Role in Skin Cancer.Int J Mol Sci. 2024 Jan 27;25(3):1582. doi: 10.3390/ijms25031582. Int J Mol Sci. 2024. PMID: 38338862 Free PMC article. Review.
-
Preferentially Expressed Antigen in Melanoma (PRAME) and the PRAME Family of Leucine-Rich Repeat Proteins.Curr Cancer Drug Targets. 2016;16(5):400-14. doi: 10.2174/1568009616666151222151818. Curr Cancer Drug Targets. 2016. PMID: 26694250 Review.
Cited by
-
The Multifaceted Role of p53 in Cancer Molecular Biology: Insights for Precision Diagnosis and Therapeutic Breakthroughs.Biomolecules. 2025 Jul 27;15(8):1088. doi: 10.3390/biom15081088. Biomolecules. 2025. PMID: 40867533 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

