Pleiotropy alleviates the fitness costs associated with resource allocation trade-offs in immune signalling networks
- PMID: 38835275
- PMCID: PMC11286129
- DOI: 10.1098/rspb.2024.0446
Pleiotropy alleviates the fitness costs associated with resource allocation trade-offs in immune signalling networks
Abstract
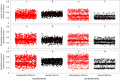
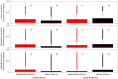
Many genes and signalling pathways within plant and animal taxa drive the expression of multiple organismal traits. This form of genetic pleiotropy instigates trade-offs among life-history traits if a mutation in the pleiotropic gene improves the fitness contribution of one trait at the expense of another. Whether or not pleiotropy gives rise to conflict among traits, however, likely depends on the resource costs and timing of trait deployment during organismal development. To investigate factors that could influence the evolutionary maintenance of pleiotropy in gene networks, we developed an agent-based model of co-evolution between parasites and hosts. Hosts comprise signalling networks that must faithfully complete a developmental programme while also defending against parasites, and trait signalling networks could be independent or share a pleiotropic component as they evolved to improve host fitness. We found that hosts with independent developmental and immune networks were significantly more fit than hosts with pleiotropic networks when traits were deployed asynchronously during development. When host genotypes directly competed against each other, however, pleiotropic hosts were victorious regardless of trait synchrony because the pleiotropic networks were more robust to parasite manipulation, potentially explaining the abundance of pleiotropy in immune systems despite its contribution to life history trade-offs.
Keywords: coevolution; evolutionary immunology; host–parasite interactions; life history trade-offs; network robustness.
Conflict of interest statement
We declare we have no competing interests.
Figures

Update of
-
Pleiotropy alleviates the fitness costs associated with resource allocation trade-offs in immune signaling networks.bioRxiv [Preprint]. 2023 Oct 10:2023.10.06.561276. doi: 10.1101/2023.10.06.561276. bioRxiv. 2023. Update in: Proc Biol Sci. 2024 Jun;291(2024):20240446. doi: 10.1098/rspb.2024.0446. PMID: 37873469 Free PMC article. Updated. Preprint.
Similar articles
-
Pleiotropy alleviates the fitness costs associated with resource allocation trade-offs in immune signaling networks.bioRxiv [Preprint]. 2023 Oct 10:2023.10.06.561276. doi: 10.1101/2023.10.06.561276. bioRxiv. 2023. Update in: Proc Biol Sci. 2024 Jun;291(2024):20240446. doi: 10.1098/rspb.2024.0446. PMID: 37873469 Free PMC article. Updated. Preprint.
-
Pleiotropy promotes the evolution of inducible immune responses in a model of host-pathogen coevolution.PLoS Comput Biol. 2023 Apr 6;19(4):e1010445. doi: 10.1371/journal.pcbi.1010445. eCollection 2023 Apr. PLoS Comput Biol. 2023. PMID: 37022993 Free PMC article.
-
Trade-Offs (and Constraints) in Organismal Biology.Physiol Biochem Zool. 2022 Jan-Feb;95(1):82-112. doi: 10.1086/717897. Physiol Biochem Zool. 2022. PMID: 34905443
-
Trade-offs, Pleiotropy, and Shared Molecular Pathways: A Unified View of Constraints on Adaptation.Integr Comp Biol. 2020 Aug 1;60(2):332-347. doi: 10.1093/icb/icaa056. Integr Comp Biol. 2020. PMID: 32483607 Review.
-
The emergence of performance trade-offs during local adaptation: insights from experimental evolution.Mol Ecol. 2017 Apr;26(7):1720-1733. doi: 10.1111/mec.13979. Epub 2017 Jan 13. Mol Ecol. 2017. PMID: 28029196 Review.
Cited by
-
Inferring the energy cost of resistance to parasitic infection and its link to a trade-off.BMC Ecol Evol. 2025 Jan 28;25(1):14. doi: 10.1186/s12862-024-02340-0. BMC Ecol Evol. 2025. PMID: 39875813 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

