Genome-wide CRISPR/Cas9 screen reveals JunB downmodulation of HIV co-receptor CXCR4
- PMID: 38835488
- PMCID: PMC11149427
- DOI: 10.3389/fmicb.2024.1342444
Genome-wide CRISPR/Cas9 screen reveals JunB downmodulation of HIV co-receptor CXCR4
Abstract
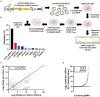
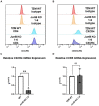
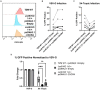
HIV-1 relies extensively on host cell machinery for replication. Identification and characterization of these host-virus interactions is vital to our understanding of viral replication and the consequences of infection in cells. Several prior screens have identified host factors important for HIV replication but with limited replication of findings, likely due to differences in experimental design and conditions. Thus, unidentified factors likely exist. To identify novel host factors required for HIV-1 infection, we performed a genome-wide CRISPR/Cas9 screen using HIV-induced cell death as a partitioning method. We created a gene knockout library in TZM-GFP reporter cells using GeCKOv2, which targets 19,050 genes, and infected the library with a lethal dose of HIV-1NL4-3. We hypothesized that cells with a knockout of a gene critical for HIV infection would survive while cells with a knockout of a non-consequential gene would undergo HIV-induced death and be lost from the population. Surviving cells were analyzed by high throughput sequencing of the integrated CRISPR/Cas9 cassette to identify the gene knockout. Of the gene targets, an overwhelming majority of the surviving cells harbored the guide sequence for the AP-1 transcription factor family protein, JunB. Upon the generation of a clonal JunB knockout cell line, we found that HIV-1NL4-3 infection was blocked in the absence of JunB. The phenotype resulted from downregulation of CXCR4, as infection levels were recovered by reintroduction of CXCR4 in JunB KO cells. Thus, JunB downmodulates CXCR4 expression in TZM-GFP cells, reducing CXCR4-tropic HIV infection.
Keywords: CRISPR/Cas9; CXCR4; HIV; JunB; host-factors.
Copyright © 2024 Schulze, Gregory, Johnson and Lange.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Simultaneous Knockout of CXCR4 and CCR5 Genes in CD4+ T Cells via CRISPR/Cas9 Confers Resistance to Both X4- and R5-Tropic Human Immunodeficiency Virus Type 1 Infection.Hum Gene Ther. 2018 Jan;29(1):51-67. doi: 10.1089/hum.2017.032. Epub 2017 Jun 9. Hum Gene Ther. 2018. PMID: 28599597
-
Genome editing of the HIV co-receptors CCR5 and CXCR4 by CRISPR-Cas9 protects CD4+ T cells from HIV-1 infection.Cell Biosci. 2017 Sep 9;7:47. doi: 10.1186/s13578-017-0174-2. eCollection 2017. Cell Biosci. 2017. PMID: 28904745 Free PMC article.
-
Genome modification of CXCR4 by Staphylococcus aureus Cas9 renders cells resistance to HIV-1 infection.Retrovirology. 2017 Nov 15;14(1):51. doi: 10.1186/s12977-017-0375-0. Retrovirology. 2017. PMID: 29141633 Free PMC article.
-
Closing the Door with CRISPR: Genome Editing of CCR5 and CXCR4 as a Potential Curative Solution for HIV.BioTech (Basel). 2022 Jul 14;11(3):25. doi: 10.3390/biotech11030025. BioTech (Basel). 2022. PMID: 35892930 Free PMC article. Review.
-
Gene Editing of HIV-1 Co-receptors to Prevent and/or Cure Virus Infection.Front Microbiol. 2018 Dec 17;9:2940. doi: 10.3389/fmicb.2018.02940. eCollection 2018. Front Microbiol. 2018. PMID: 30619107 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

