Gene-deficient mouse model established by CRISPR/Cas9 system reveals 15 reproductive organ-enriched genes dispensable for male fertility
- PMID: 38835510
- PMCID: PMC11148293
- DOI: 10.3389/fcell.2024.1411162
Gene-deficient mouse model established by CRISPR/Cas9 system reveals 15 reproductive organ-enriched genes dispensable for male fertility
Abstract
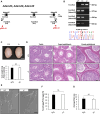
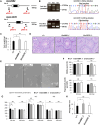
Since the advent of gene-targeting technology in embryonic stem cells, mice have become a primary model organism for investigating human gene function due to the striking genomic similarities between the two species. With the introduction of the CRISPR/Cas9 system for genome editing in mice, the pace of loss-of-function analysis has accelerated significantly. This has led to the identification of numerous genes that play crucial roles in male reproductive processes, including meiosis, chromatin condensation, flagellum formation in the testis, sperm maturation in the epididymis, and fertilization in the oviduct. Despite the advancements, the functions of many genes, particularly those enriched in male reproductive tissues, remain largely unknown. In our study, we focused on 15 genes and generated 13 gene-deficient mice [4933411K16Rik, Adam triple (Adam20, Adam25, and Adam39), BC048671, Cfap68, Gm4846, Gm4984, Gm13570, Nt5c1b, Ppp1r42, Saxo4, Sh3d21, Spz1, and Tektl1] to elucidate their roles in male fertility. Surprisingly, all 13 gene-deficient mice exhibited normal fertility in natural breeding experiments, indicating that these genes are not essential for male fertility. These findings have important implications as they may help prevent other research laboratories from duplicating efforts to generate knockout mice for genes that do not demonstrate an apparent phenotype related to male fertility. By shedding light on the dispensability of these genes, our study contributes to a more efficient allocation of research resources in the exploration of male reproductive biology.
Keywords: CRISPR/Cas9; knockout mice; male infertility; spermatozoa; testis.
Copyright © 2024 Nguyen, Tokuhiro, Shimada, Wang, Mashiko, Tonai, Kiyozumi and Ikawa.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
CRISPR/Cas9-mediated genome editing reveals 30 testis-enriched genes dispensable for male fertility in mice†.Biol Reprod. 2019 Aug 1;101(2):501-511. doi: 10.1093/biolre/ioz103. Biol Reprod. 2019. PMID: 31201419 Free PMC article.
-
CRISPR/Cas9-mediated genome editing reveals 12 testis-enriched genes dispensable for male fertility in mice.Asian J Androl. 2022 May-Jun;24(3):266-272. doi: 10.4103/aja.aja_63_21. Asian J Androl. 2022. PMID: 34290169 Free PMC article.
-
Genome engineering uncovers 54 evolutionarily conserved and testis-enriched genes that are not required for male fertility in mice.Proc Natl Acad Sci U S A. 2016 Jul 12;113(28):7704-10. doi: 10.1073/pnas.1608458113. Epub 2016 Jun 29. Proc Natl Acad Sci U S A. 2016. PMID: 27357688 Free PMC article.
-
Revolutionizing male fertility factor research in mice by using the genome editing tool CRISPR/Cas9.Reprod Med Biol. 2017 Oct 14;17(1):3-10. doi: 10.1002/rmb2.12067. eCollection 2018 Jan. Reprod Med Biol. 2017. PMID: 29371815 Free PMC article. Review.
-
Disrupting the male germ line to find infertility and contraception targets.Ann Endocrinol (Paris). 2014 May;75(2):101-8. doi: 10.1016/j.ando.2014.04.006. Epub 2014 Apr 30. Ann Endocrinol (Paris). 2014. PMID: 24793995 Review.
Cited by
-
Long-read RNA-sequencing reveals transcript-specific regulation in human-derived cortical neurons.Open Biol. 2025 Jul;15(7):250200. doi: 10.1098/rsob.250200. Epub 2025 Jul 30. Open Biol. 2025. PMID: 40735840 Free PMC article.
-
Brief biology and pathophysiology of Tekt bundles.Cell Adh Migr. 2025 Dec;19(1):2465421. doi: 10.1080/19336918.2025.2465421. Epub 2025 Feb 13. Cell Adh Migr. 2025. PMID: 39949046 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

