Identification, molecular evolution, codon bias, and expansion analysis of NLP transcription factor family in foxtail millet (Setaria italica L.) and closely related crops
- PMID: 38836039
- PMCID: PMC11148446
- DOI: 10.3389/fgene.2024.1395224
Identification, molecular evolution, codon bias, and expansion analysis of NLP transcription factor family in foxtail millet (Setaria italica L.) and closely related crops
Abstract
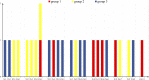
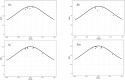
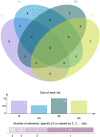
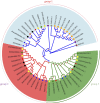
The NODULE-INCEPTION-like protein (NLP) family is a plant-specific transcription factor (TF) family involved in nitrate transport and assimilation in plants, which are essential for improving plant nitrogen use efficiency. Currently, the molecular nature and evolutionary trajectory of NLP genes in the C4 model crop foxtail millet are unknown. Therefore, we performed a comprehensive analysis of NLP and molecular evolution in foxtail millet by scanning the genomes of foxtail millet and representative species of the plant kingdom. We identified seven NLP genes in the foxtail millet genome, all of which are individually and separately distributed on different chromosomes. They were not structurally identical to each other and were mainly expressed on root tissues. We unearthed two key genes (Si5G004100.1 and Si6G248300.1) with a variety of excellent characteristics. Regarding its molecular evolution, we found that NLP genes in Gramineae mainly underwent dispersed duplication, but maize NLP genes were mainly generated via WGD events. Other factors such as base mutations and natural selection have combined to promote the evolution of NLP genes. Intriguingly, the family in plants showed a gradual expansion during evolution with more duplications than losses, contrary to most gene families. In conclusion, this study advances the use of NLP genetic resources and the understanding of molecular evolution in cereals.
Keywords: NODULE-INCEPTION-like protein; codon bias; foxtail millet; molecular evolution; structure; transcription factor.
Copyright © 2024 Chen, Liu, Chen, Ji, Cui, Ge and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Genome-Wide Identification of NLP Gene Families and Haplotype Analysis of SiNLP2 in Foxtail Millet (Setaria italica).Int J Mol Sci. 2024 Dec 2;25(23):12938. doi: 10.3390/ijms252312938. Int J Mol Sci. 2024. PMID: 39684649 Free PMC article.
-
Genome-wide identification and expression analysis of the bHLH transcription factor family and its response to abiotic stress in foxtail millet (Setaria italica L.).BMC Genomics. 2021 Oct 30;22(1):778. doi: 10.1186/s12864-021-08095-y. BMC Genomics. 2021. PMID: 34717536 Free PMC article.
-
Identification and molecular characterization of MYB Transcription Factor Superfamily in C4 model plant foxtail millet (Setaria italica L.).PLoS One. 2014 Oct 3;9(10):e109920. doi: 10.1371/journal.pone.0109920. eCollection 2014. PLoS One. 2014. PMID: 25279462 Free PMC article.
-
Adaptation of Foxtail Millet (Setaria italica L.) to Abiotic Stresses: A Special Perspective of Responses to Nitrogen and Phosphate Limitations.Front Plant Sci. 2020 Feb 28;11:187. doi: 10.3389/fpls.2020.00187. eCollection 2020. Front Plant Sci. 2020. PMID: 32184798 Free PMC article. Review.
-
Foxtail millet: a model crop for genetic and genomic studies in bioenergy grasses.Crit Rev Biotechnol. 2013 Sep;33(3):328-43. doi: 10.3109/07388551.2012.716809. Epub 2012 Sep 18. Crit Rev Biotechnol. 2013. PMID: 22985089 Review.
Cited by
-
Identification and expression characteristics of NIN-like protein (NLP) gene family in cucumber plant (Cucumis sativus L.).BMC Plant Biol. 2025 Aug 30;25(1):1163. doi: 10.1186/s12870-025-07203-4. BMC Plant Biol. 2025. PMID: 40885889 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

