Untargeted metabolomics and metagenomics reveal signatures for intramammary ceftiofur treatment and lactation stage in the cattle hindgut
- PMID: 38836107
- PMCID: PMC11148447
- DOI: 10.3389/fmolb.2024.1364637
Untargeted metabolomics and metagenomics reveal signatures for intramammary ceftiofur treatment and lactation stage in the cattle hindgut
Abstract
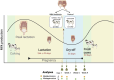
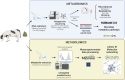
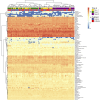
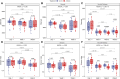
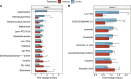
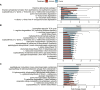
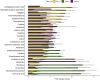
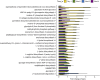
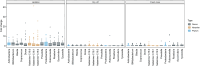
The gut microbiota in cattle is essential for protein, energy, and vitamin production and hence, microbiota perturbations can affect cattle performance. This study evaluated the effect of intramammary (IMM) ceftiofur treatment and lactation stage on the functional gut microbiome and metabolome. Forty dairy cows were enrolled at dry-off. Half received IMM ceftiofur and a non-antibiotic teat sealant containing bismuth subnitrate (cases), while the other half received the teat sealant (controls). Fecal samples were collected before treatment at dry off, during the dry period (weeks 1 and 5) and the first week after calving (week 9). Shotgun metagenomic sequencing was applied to predict microbial metabolic pathways whereas untargeted metabolomics was used identify polar and nonpolar metabolites. Compared to controls, long-term changes were observed in the cows given ceftiofur, including a lower abundance of microbial pathways linked to energy production, amino acid biosynthesis, and other vital molecules. The metabolome of treated cows had elevated levels of stachyose, phosphatidylethanolamine diacylglycerol (PE-DAG), and inosine a week after the IMM ceftiofur application, indicating alterations in microbial fermentation, lipid metabolism, energy, and cellular signaling. Differences were also observed by sampling, with cows in late lactation having more diverse metabolic pathways and a unique metabolome containing higher levels of histamine and histamine-producing bacteria. These data illustrate how IMM ceftiofur treatment can alter the functionality of the hindgut metabolome and microbiome. Understanding how antibiotics and lactation stages, which are each characterized by unique diets and physiology, impact the function of resident microbes is critical to define normal gut function in dairy cattle.
Keywords: antibiotic use; ceftiofur; gut microbiome; metabolomics; metagenomics.
Copyright © 2024 Vasco, Hansen, Schilmiller, Bowcutt, Carbonell, Ruegg, Quinn, Zhang and Manning.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures

Similar articles
-
Persistent effects of intramammary ceftiofur treatment on the gut microbiome and antibiotic resistance in dairy cattle.Anim Microbiome. 2023 Nov 9;5(1):56. doi: 10.1186/s42523-023-00274-4. Anim Microbiome. 2023. PMID: 37946266 Free PMC article.
-
Longitudinal characterization of the metabolome of dairy cows transitioning from one lactation to the next: Investigations in blood serum.J Dairy Sci. 2024 Feb;107(2):1263-1285. doi: 10.3168/jds.2023-23841. Epub 2023 Sep 29. J Dairy Sci. 2024. PMID: 37777004
-
The effect of internal teat sealant products (Teatseal and Orbeseal) on intramammary infection, clinical mastitis, and somatic cell counts in lactating dairy cows: a meta-analysis.J Dairy Sci. 2013;96(11):6915-6931. doi: 10.3168/jds.2013-6544. Epub 2013 Sep 18. J Dairy Sci. 2013. PMID: 24054298
-
Effect of dry period length on the effect of an intramammary teat sealant on the risk of mastitis in cattle treated with antibiotics at drying off.N Z Vet J. 2014 Jul;62(4):214-20. doi: 10.1080/00480169.2013.879689. Epub 2014 Feb 28. N Z Vet J. 2014. PMID: 24472043 Clinical Trial.
-
Non-antimicrobial approaches at drying-off for treating and preventing intramammary infections in dairy cows. Part 1. Meta-analyses of efficacy of using an internal teat sealant without a concomitant antimicrobial treatment.Anim Health Res Rev. 2019 Jun;20(1):86-97. doi: 10.1017/S1466252319000070. Epub 2019 Nov 26. Anim Health Res Rev. 2019. PMID: 31895023
References
Grants and funding
LinkOut - more resources
Full Text Sources

