The novel lnc-HZ12 suppresses autophagy degradation of BBC3 by preventing its interactions with HSPA8 to induce trophoblast cell apoptosis
- PMID: 38836496
- PMCID: PMC11423690
- DOI: 10.1080/15548627.2024.2362122
The novel lnc-HZ12 suppresses autophagy degradation of BBC3 by preventing its interactions with HSPA8 to induce trophoblast cell apoptosis
Abstract
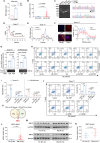
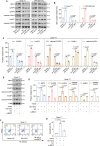
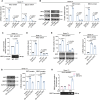
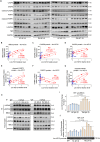
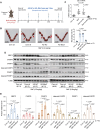
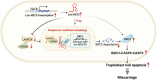
Abnormal expression of long non-coding RNAs (lncRNAs) is associated with the dysfunctions of human trophoblast cells and the occurrence of miscarriage (abnormal early embryo loss). BBC3/PUMA (BCL2 binding component 3) plays significant roles in regulation of cell apoptosis. However, whether specific lncRNAs might regulate BBC3 in trophoblast cells and further induce apoptosis and miscarriage remains completely unclear. Through screening, we identified a novel lnc-HZ12, which was significantly highly expressed in villous tissues of recurrent miscarriage (RM) patients relative to their healthy control (HC) group. Lnc-HZ12 suppressed chaperone-mediated autophagy (CMA) degradation of BBC3, promoted trophoblast cell apoptosis, and was associated with miscarriage. In mechanism, lnc-HZ12 downregulated the expression levels of chaperone molecules HSPA8 and LAMP2A in trophoblast cells. Meanwhile, lnc-HZ12 (mainly lnc-HZ12-SO2 region in F2 fragment) and HSPA8 competitively bound with the 169RVLYNL174 patch on BBC3, which prevented BBC3 from interactions with HSPA8 and impaired the formation of BBC3-HSPA8-LAMP2A complex for CMA degradation of BBC3. Thus, lnc-HZ12 upregulated the BBC3-CASP9-CASP3 pathway and induced trophoblast cell apoptosis. In villous tissues, lnc-HZ12 was highly expressed, CMA degradation of BBC3 was suppressed, and the apoptosis levels were higher in RM vs HC villous tissues, all of which were associated with miscarriage. Interestingly, knockdown of murine Bbc3 could efficiently suppress placental apoptosis and alleviate miscarriage in a mouse miscarriage model. Taken together, our results indicated that lnc-HZ12 and BBC3 played important roles in trophoblast cell apoptosis and miscarriage and might act as attractive targets for miscarriage treatment.Abbreviation: 7-AAD: 7-aminoactinomycin D; BaP: benzopyrene; BBC3/PUMA: BCL2 binding component 3; ChIP: chromatin immunoprecipitation; CHX: cycloheximide; CMA: chaperone-mediated autophagy; CQ: chloroquine; DMSO: dimethyl sulfoxide; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; HC: healthy control; HSPA8: heat shock protein family A (Hsp70) member 8; IP: immunoprecipitation; LAMP2A: lysosomal associated membrane protein 2; LncRNA: long non-coding RNA; mRNA: messenger RNA; MT: mutant-type; NC: negative control; NSO: nonspecific oligonucleotide; PARP1: poly(ADP-ribose) polymerase 1; RIP: RNA immunoprecipitation; RM: recurrent miscarriage; TBP: TATA-box binding protein; WT: wild-type.
Keywords: Apoptosis; BBC3/PUMA-CASP9-CASP3; chaperone-mediated autophagy (CMA); human trophoblast cells; lncRNA or lnc-HZ12; recurrent miscarriage villous tissues.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



References
-
- Umehara N, Tanaka T.. The incidence of various antiphospholipid antibodies, measured by commercial-based laboratory, with recurrent spontenous abortion and the impact of their profiles on reproductive outcome with active anticoagulant therapy. ISRN Obstet Gynecol. 2012;2012(3):819356. doi: 10.5402/2012/819356 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
