Lamotrigine-mediated rescue of RsgA-deficient Escherichia coli reveals another role of IF2 in ribosome biogenesis
- PMID: 38837341
- PMCID: PMC11270870
- DOI: 10.1128/jb.00119-24
Lamotrigine-mediated rescue of RsgA-deficient Escherichia coli reveals another role of IF2 in ribosome biogenesis
Abstract
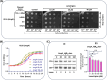
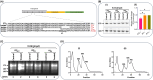
RsgA (small ribosomal subunit, 30S, GTPase), a late-stage biogenesis factor, releases RbfA from 30S-RbfA complex. Escherichia coli ΔrsgA (deleted for rsgA) shows a slow growth phenotype and an increased accumulation of 17S rRNA (precursor of 16S rRNA) and the ribosomal subunits. Here, we show that the rescue of the ΔrsgA strain by multicopy infB (IF2) is enhanced by simultaneous overexpression of initiator tRNA (i-tRNA), suggesting a role of initiation complex formation in growth rescue. The synergistic effect of IF2/i-tRNA is accompanied by increased processing of 17S rRNA (to 16S), and protection of the 16S rRNA 3'-minor domain. Importantly, we show that an IF2-binding anticonvulsant drug, lamotrigine (Ltg), also rescues the ΔrsgA strain growth. The rescue is accompanied by increased processing of 17S rRNA, protection of the 3'-minor domain of 16S rRNA, and increased 70S ribosomes in polysome profiles. However, Ltg becomes inhibitory to the ΔrsgA strain whose growth was already rescued by an L83R mutation in rbfA. Interestingly, like wild-type infB, overproduction of LtgRinfB alleles (having indel mutations in their domain II) also rescues the ΔrsgA strain (independent of Ltg). Our observations suggest the dual role of IF2 in rescuing the ΔrsgA strain. First, together with i-tRNA, IF2 facilitates the final steps of processing of 17S rRNA. Second, a conformer of IF2 functionally compensates for RsgA, albeit poorly, during 30S biogenesis.
Importance: RsgA is a late-stage ribosome biogenesis factor. Earlier, infB (IF2) was isolated as a multicopy suppressor of the Escherichia coli ΔrsgA strain. How IF2 rescued the strain growth remained unclear. This study reveals that (i) the multicopy infB-mediated growth rescue of E. coli ΔrsgA and the processing of 17S precursor to 16S rRNA in the strain are enhanced upon simultaneous overexpression of initiator tRNA and (ii) a conformer of IF2, whose occurrence increases when IF2 is overproduced or when E. coli ΔrsgA is treated with Ltg (an anticonvulsant drug that binds to domain II of IF2), compensates for the function of RsgA. Thus, this study reveals yet another role of IF2 in ribosome biogenesis.
Keywords: 3GC base pairs; RbfA; initiation factor 2; initiator tRNA; rRNA maturation; ribosome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Lamotrigine compromises the fidelity of initiator tRNA recruitment to the ribosomal P-site by IF2 and the RbfA release from 30S ribosomes in Escherichia coli.RNA Biol. 2023 Jan;20(1):681-692. doi: 10.1080/15476286.2023.2253395. RNA Biol. 2023. PMID: 37676049 Free PMC article.
-
RsgA releases RbfA from 30S ribosome during a late stage of ribosome biosynthesis.EMBO J. 2011 Jan 5;30(1):104-14. doi: 10.1038/emboj.2010.291. Epub 2010 Nov 23. EMBO J. 2011. PMID: 21102555 Free PMC article.
-
Conserved GTPase LepA (Elongation Factor 4) functions in biogenesis of the 30S subunit of the 70S ribosome.Proc Natl Acad Sci U S A. 2017 Jan 31;114(5):980-985. doi: 10.1073/pnas.1613665114. Epub 2017 Jan 17. Proc Natl Acad Sci U S A. 2017. PMID: 28096346 Free PMC article.
-
Roles of elusive translational GTPases come to light and inform on the process of ribosome biogenesis in bacteria.Mol Microbiol. 2018 Feb;107(4):445-454. doi: 10.1111/mmi.13895. Epub 2017 Dec 29. Mol Microbiol. 2018. PMID: 29235176 Free PMC article. Review.
-
Structural insights into cell cycle control by essential GTPase Era.Postepy Biochem. 2016;62(3):335-342. Postepy Biochem. 2016. PMID: 28132488 Free PMC article. Review.
Cited by
-
Role of the sarcin-ricin loop of 23S rRNA in biogenesis of the 50S ribosomal subunit.RNA. 2025 Mar 18;31(4):585-599. doi: 10.1261/rna.080335.124. RNA. 2025. PMID: 39875174 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

