Drug-target interaction predictions with multi-view similarity network fusion strategy and deep interactive attention mechanism
- PMID: 38837345
- PMCID: PMC11164831
- DOI: 10.1093/bioinformatics/btae346
Drug-target interaction predictions with multi-view similarity network fusion strategy and deep interactive attention mechanism
Abstract
Motivation: Accurately identifying the drug-target interactions (DTIs) is one of the crucial steps in the drug discovery and drug repositioning process. Currently, many computational-based models have already been proposed for DTI prediction and achieved some significant improvement. However, these approaches pay little attention to fuse the multi-view similarity networks related to drugs and targets in an appropriate way. Besides, how to fully incorporate the known interaction relationships to accurately represent drugs and targets is not well investigated. Therefore, there is still a need to improve the accuracy of DTI prediction models.
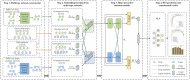
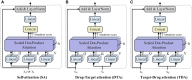
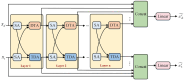
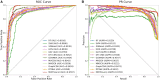
Results: In this study, we propose a novel approach that employs Multi-view similarity network fusion strategy and deep Interactive attention mechanism to predict Drug-Target Interactions (MIDTI). First, MIDTI constructs multi-view similarity networks of drugs and targets with their diverse information and integrates these similarity networks effectively in an unsupervised manner. Then, MIDTI obtains the embeddings of drugs and targets from multi-type networks simultaneously. After that, MIDTI adopts the deep interactive attention mechanism to further learn their discriminative embeddings comprehensively with the known DTI relationships. Finally, we feed the learned representations of drugs and targets to the multilayer perceptron model and predict the underlying interactions. Extensive results indicate that MIDTI significantly outperforms other baseline methods on the DTI prediction task. The results of the ablation experiments also confirm the effectiveness of the attention mechanism in the multi-view similarity network fusion strategy and the deep interactive attention mechanism.
Availability and implementation: https://github.com/XuLew/MIDTI.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures


Similar articles
-
MHADTI: predicting drug-target interactions via multiview heterogeneous information network embedding with hierarchical attention mechanisms.Brief Bioinform. 2022 Nov 19;23(6):bbac434. doi: 10.1093/bib/bbac434. Brief Bioinform. 2022. PMID: 36242566
-
H2GnnDTI: hierarchical heterogeneous graph neural networks for drug-target interaction prediction.Bioinformatics. 2025 Mar 29;41(4):btaf117. doi: 10.1093/bioinformatics/btaf117. Bioinformatics. 2025. PMID: 40097269 Free PMC article.
-
MiRAGE-DTI: A novel approach for drug-target interaction prediction by integrating drug and target similarity metrics.Comput Biol Med. 2025 Jun;192(Pt B):110249. doi: 10.1016/j.compbiomed.2025.110249. Epub 2025 May 12. Comput Biol Med. 2025. PMID: 40359678
-
Open-source chemogenomic data-driven algorithms for predicting drug-target interactions.Brief Bioinform. 2019 Jul 19;20(4):1465-1474. doi: 10.1093/bib/bby010. Brief Bioinform. 2019. PMID: 29420684 Free PMC article. Review.
-
Research progress on Drug-Target Interactions in the last five years.Anal Biochem. 2025 Feb;697:115691. doi: 10.1016/j.ab.2024.115691. Epub 2024 Oct 23. Anal Biochem. 2025. PMID: 39455038 Review.
Cited by
-
Dynamic category-sensitive hypergraph inferring and homo-heterogeneous neighbor feature learning for drug-related microbe prediction.Bioinformatics. 2024 Sep 2;40(9):btae562. doi: 10.1093/bioinformatics/btae562. Bioinformatics. 2024. PMID: 39292557 Free PMC article.
-
DTI-RME: a robust and multi-kernel ensemble approach for drug-target interaction prediction.BMC Biol. 2025 Jul 28;23(1):225. doi: 10.1186/s12915-025-02340-6. BMC Biol. 2025. PMID: 40717088 Free PMC article.
-
Harnessing artificial intelligence to identify Bufalin as a molecular glue degrader of estrogen receptor alpha.Nat Commun. 2025 Aug 22;16(1):7854. doi: 10.1038/s41467-025-62288-7. Nat Commun. 2025. PMID: 40846852 Free PMC article.
-
DTI-MHAPR: optimized drug-target interaction prediction via PCA-enhanced features and heterogeneous graph attention networks.BMC Bioinformatics. 2025 Jan 13;26(1):11. doi: 10.1186/s12859-024-06021-z. BMC Bioinformatics. 2025. PMID: 39800678 Free PMC article.
References
-
- Chang C-C, Lin C-J.. Libsvm: a library for support vector machines. ACM Trans Intell Syst Technol 2011;2:1–27.
-
- Chen T, Guestrin C. Xgboost: A scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 2016, 785–94.
-
- Dai Q, Liu Z, Wang Z. et al. Graphcda: a hybrid graph representation learning framework based on gcn and gat for predicting disease-associated circrnas. Brief Bioinform 2022;23:bbac379. - PubMed
-
- Ding Y, Tang J, Guo F. et al. Identification of drug–target interactions via multiple kernel-based triple collaborative matrix factorization. Brief Bioinform 2022;23:bbab582. - PubMed

