Human cytomegalovirus glycoprotein variants governing viral tropism and syncytium formation in epithelial cells and macrophages
- PMID: 38837351
- PMCID: PMC11265420
- DOI: 10.1128/jvi.00293-24
Human cytomegalovirus glycoprotein variants governing viral tropism and syncytium formation in epithelial cells and macrophages
Abstract
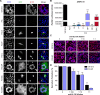
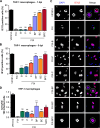
Human cytomegalovirus (HCMV) displays a broad cell tropism, and the infection of biologically relevant cells such as epithelial, endothelial, and hematopoietic cells supports viral transmission, systemic spread, and pathogenesis in the human host. HCMV strains differ in their ability to infect and replicate in these cell types, but the genetic basis of these differences has remained incompletely understood. In this study, we investigated HCMV strain VR1814, which is highly infectious for epithelial cells and macrophages and induces cell-cell fusion in both cell types. A VR1814-derived bacterial artificial chromosome (BAC) clone, FIX-BAC, was generated many years ago but has fallen out of favor because of its modest infectivity. By sequence comparison and genetic engineering of FIX, we demonstrate that the high infectivity of VR1814 and its ability to induce syncytium formation in epithelial cells and macrophages depends on VR1814-specific variants of the envelope glycoproteins gB, UL128, and UL130. We also show that UL130-neutralizing antibodies inhibit syncytium formation, and a FIX-specific mutation in UL130 is responsible for its low infectivity by reducing the amount of the pentameric glycoprotein complex in viral particles. Moreover, we found that a VR1814-specific mutation in US28 further increases viral infectivity in macrophages, possibly by promoting lytic rather than latent infection of these cells. Our findings show that variants of gB and the pentameric complex are major determinants of infectivity and syncytium formation in epithelial cells and macrophages. Furthermore, the VR1814-adjusted FIX strains can serve as valuable tools to study HCMV infection of myeloid cells.IMPORTANCEHuman cytomegalovirus (HCMV) is a major cause of morbidity and mortality in transplant patients and the leading cause of congenital infections. HCMV infects various cell types, including epithelial cells and macrophages, and some strains induce the fusion of neighboring cells, leading to the formation of large multinucleated cells called syncytia. This process may limit the exposure of the virus to host immune factors and affect pathogenicity. However, the reason why some HCMV strains exhibit a broader cell tropism and why some induce cell fusion more than others is not well understood. We compared two closely related HCMV strains and provided evidence that small differences in viral envelope glycoproteins can massively increase or decrease the virus infectivity and its ability to induce syncytium formation. The results of the study suggest that natural strain variations may influence HCMV infection and pathogenesis in humans.
Keywords: cell fusion; entry; glycoproteins; human cytomegalovirus; infectivity; strain variation; syncytium.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Mocarski ES, Shenk T, Griffiths PD, Pass RF. 2013. Cytomegaloviruses. In Knipe DM, Howley PM (ed), Field’s virology, 6th ed. Vol. 1. Lippincott Williams & Wilkin, Philadelphia.
-
- Rawlinson WD, Boppana SB, Fowler KB, Kimberlin DW, Lazzarotto T, Alain S, Daly K, Doutré S, Gibson L, Giles ML, Greenlee J, Hamilton ST, Harrison GJ, Hui L, Jones CA, Palasanthiran P, Schleiss MR, Shand AW, van Zuylen WJ. 2017. Congenital cytomegalovirus infection in pregnancy and the neonate: consensus recommendations for prevention, diagnosis, and therapy. Lancet Infect Dis 17:e177–e188. doi:10.1016/S1473-3099(17)30143-3 - DOI - PubMed
-
- Sinzger C, Grefte A, Plachter B, Gouw AS, The TH, Jahn G. 1995. Fibroblasts, epithelial cells, endothelial cells and smooth muscle cells are major targets of human cytomegalovirus infection in lung and gastrointestinal tissues. J Gen Virol 76 (Pt 4):741–750. doi:10.1099/0022-1317-76-4-741 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

