Inflammatory caspase substrate specificities
- PMID: 38837391
- PMCID: PMC11253702
- DOI: 10.1128/mbio.02975-23
Inflammatory caspase substrate specificities
Abstract
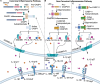
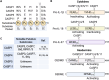
Caspases are a family of cysteine proteases that act as molecular scissors to cleave substrates and regulate biological processes such as programmed cell death and inflammation. Extensive efforts have been made to identify caspase substrates and to determine factors that dictate substrate specificity. Thousands of putative substrates have been identified for caspases that regulate an immunologically silent type of cell death known as apoptosis, but less is known about substrates of the inflammatory caspases that regulate an immunostimulatory type of cell death called pyroptosis. Furthermore, much of our understanding of caspase substrate specificities is derived from work done with peptide substrates, which do not often translate to native protein substrates. Our knowledge of inflammatory caspase biology and substrates has recently expanded and here, we discuss the recent advances in our understanding of caspase substrate specificities, with a focus on inflammatory caspases. We highlight new substrates that have been discovered and discuss the factors that engender specificity. Recent evidence suggests that inflammatory caspases likely utilize two binding interfaces to recognize and process substrates, the active site and a conserved exosite.
Keywords: IL-18; IL-1β; caspase substrates; caspase-1; caspase-11; caspase-4; caspase-5; cytokines; inflammasomes; inflammatory caspases; pyroptosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Chemical Tools Based on the Tetrapeptide Sequence of IL-18 Reveals Shared Specificities between Inflammatory and Apoptotic Initiator Caspases.bioRxiv [Preprint]. 2025 Feb 27:2025.02.23.639785. doi: 10.1101/2025.02.23.639785. bioRxiv. 2025. PMID: 40060427 Free PMC article. Preprint.
-
Extensive peptide and natural protein substrate screens reveal that mouse caspase-11 has much narrower substrate specificity than caspase-1.J Biol Chem. 2018 May 4;293(18):7058-7067. doi: 10.1074/jbc.RA117.001329. Epub 2018 Feb 6. J Biol Chem. 2018. PMID: 29414788 Free PMC article.
-
Caspases in Cell Death, Inflammation, and Disease.Immunity. 2019 Jun 18;50(6):1352-1364. doi: 10.1016/j.immuni.2019.05.020. Immunity. 2019. PMID: 31216460 Free PMC article. Review.
-
Great balls of fire: activation and signalling of inflammatory caspases.Biochem Soc Trans. 2021 Jun 30;49(3):1311-1324. doi: 10.1042/BST20200986. Biochem Soc Trans. 2021. PMID: 34060593 Free PMC article. Review.
-
Extended subsite profiling of the pyroptosis effector protein gasdermin D reveals a region recognized by inflammatory caspase-11.J Biol Chem. 2020 Aug 7;295(32):11292-11302. doi: 10.1074/jbc.RA120.014259. Epub 2020 Jun 18. J Biol Chem. 2020. PMID: 32554464 Free PMC article.
Cited by
-
The molecular mechanisms, roles, and potential applications of PANoptosis in cancer treatment.Front Immunol. 2025 Apr 29;16:1550800. doi: 10.3389/fimmu.2025.1550800. eCollection 2025. Front Immunol. 2025. PMID: 40364845 Free PMC article. Review.
-
Chemical Tools Based on the Tetrapeptide Sequence of IL-18 Reveals Shared Specificities between Inflammatory and Apoptotic Initiator Caspases.bioRxiv [Preprint]. 2025 Feb 27:2025.02.23.639785. doi: 10.1101/2025.02.23.639785. bioRxiv. 2025. PMID: 40060427 Free PMC article. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
- Penn Provost Postdoctoral Fellow/University of Pennsylvania Perelman School of Medicine
- 1054907/Burroughs Wellcome Fund (BWF)
- Martin and Pamela Winter Infectious Disease Fellowship./University of Pennsylvania IIZD
- EE Just Early Career Award/United Negro College Fund (UNCF)
- R00 AI148598/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources

