Targeting Rab7-Rilp Mediated Microlipophagy Alleviates Lipid Toxicity in Diabetic Cardiomyopathy
- PMID: 38837607
- PMCID: PMC11304244
- DOI: 10.1002/advs.202401676
Targeting Rab7-Rilp Mediated Microlipophagy Alleviates Lipid Toxicity in Diabetic Cardiomyopathy
Abstract
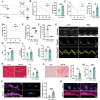
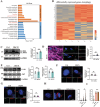
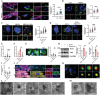
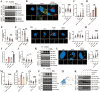
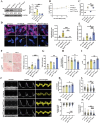
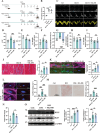
Diabetic cardiomyopathy (DbCM) is characterized by diastolic dysfunction, which progresses into heart failure and aberrant electrophysiology in diabetic patients. Dyslipidemia in type 2 diabetic patients leads to the accumulation of lipid droplets (LDs) in cardiomyocytes and results in lipid toxicity which has been suggested to drive DbCM. It is aimed to explore potential pathways that may boost LDs degradation in DbCM and restore cardiac function. LDs accumulation resulted in an increase in lipid toxicity in DbCM hearts is confirmed. Microlipophagy pathway, rather than traditional macrolipophagy, is activated in DbCM hearts. RNA-Seq data and Rab7-CKO mice implicate that Rab7 is a major modulator of the microlipophagy pathway. Mechanistically, Rab7 is phosphorylated at Tyrosine 183, which allows the recruitment of Rab-interacting lysosome protein (Rilp) to proceed LDs degradation by lysosome. Treating DbCM mice with Rab7 activator ML-098 enhanced Rilp level and rescued the observed cardiac dysfunction. Overall, Rab7-Rilp-mediated microlipophagy may be a promising target in the treatment of lipid toxicity in DbCM is suggested.
Keywords: diabetic cardiomyopathy; lipid toxicity; lipophagy.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Rubler S., Dlugash J., Yuceoglu Y. Z., Kumral T., Branwood A. W., Grishman A., Am J Cardiol 1972, 30, 595. - PubMed
-
- Seferović P. M., Paulus W. J., Eur. Heart J. 2015, 36, 1718. - PubMed
-
- Liu J. E., Palmieri V., Roman M. J., Bella J. N., Fabsitz R., Howard B. V., Welty T. K., Lee E. T., Devereux R. B., J. Am. Coll. Cardiol. 2001, 37, 1943. - PubMed
-
- Segar M. W., Khan M. S., Patel K. V., Butler J., Tang W. H. W., Vaduganathan M., Lam C. S. P., Verma S., McGuire D. K., Pandey A., J. Am. Coll. Cardiol. 2021, 78, 1587. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
