MechanoProDB: a web-based database for exploring the mechanical properties of proteins
- PMID: 38837788
- PMCID: PMC11152175
- DOI: 10.1093/database/baae047
MechanoProDB: a web-based database for exploring the mechanical properties of proteins
Abstract
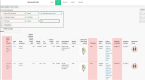
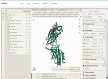
The mechanical stability of proteins is crucial for biological processes. To understand the mechanical functions of proteins, it is important to know the protein structure and mechanical properties. Protein mechanics is usually investigated through force spectroscopy experiments and simulations that probe the forces required to unfold the protein of interest. While there is a wealth of data in the literature on force spectroscopy experiments and steered molecular dynamics simulations of forced protein unfolding, this information is spread and difficult to access by non-experts. Here, we introduce MechanoProDB, a novel web-based database resource for collecting and mining data obtained from experimental and computational works. MechanoProDB provides a curated repository for a wide range of proteins, including muscle proteins, adhesion molecules and membrane proteins. The database incorporates relevant parameters that provide insights into the mechanical stability of proteins and their conformational stability such as the unfolding forces, energy landscape parameters and contour lengths of unfolding steps. Additionally, it provides intuitive annotations of the unfolding pathways of each protein, allowing users to explore the individual steps during mechanical unfolding. The user-friendly interface of MechanoProDB allows researchers to efficiently navigate, search and download data pertaining to specific protein folds or experimental conditions. Users can visualize protein structures using interactive tools integrated within the database, such as Mol*, and plot available data through integrated plotting tools. To ensure data quality and reliability, we have carefully manually verified and curated the data currently available on MechanoProDB. Furthermore, the database also features an interface that enables users to contribute new data and annotations, promoting community-driven comprehensiveness. The freely available MechanoProDB aims to streamline and accelerate research in the field of mechanobiology and biophysics by offering a unique platform for data sharing and analysis. MechanoProDB is freely available at https://mechanoprodb.ibdm.univ-amu.fr.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures




Similar articles
-
PPInterface: A Comprehensive Dataset of 3D Protein-Protein Interface Structures.J Mol Biol. 2024 Sep 1;436(17):168686. doi: 10.1016/j.jmb.2024.168686. Epub 2024 Jun 25. J Mol Biol. 2024. PMID: 38936693
-
ProteomeScout: a repository and analysis resource for post-translational modifications and proteins.Nucleic Acids Res. 2015 Jan;43(Database issue):D521-30. doi: 10.1093/nar/gku1154. Epub 2014 Nov 20. Nucleic Acids Res. 2015. PMID: 25414335 Free PMC article.
-
PDTD: a web-accessible protein database for drug target identification.BMC Bioinformatics. 2008 Feb 19;9:104. doi: 10.1186/1471-2105-9-104. BMC Bioinformatics. 2008. PMID: 18282303 Free PMC article.
-
IIS--Integrated Interactome System: a web-based platform for the annotation, analysis and visualization of protein-metabolite-gene-drug interactions by integrating a variety of data sources and tools.PLoS One. 2014 Jun 20;9(6):e100385. doi: 10.1371/journal.pone.0100385. eCollection 2014. PLoS One. 2014. PMID: 24949626 Free PMC article.
-
Experimental and theoretical studies of mechanical unfolding of different proteins.Biochemistry (Mosc). 2013 Nov;78(11):1216-27. doi: 10.1134/S0006297913110023. Biochemistry (Mosc). 2013. PMID: 24460936 Review.
Cited by
-
MechanoBase: a comprehensive database for the mechanics of tissues and cells.Database (Oxford). 2024 May 28;2024:baae040. doi: 10.1093/database/baae040. Database (Oxford). 2024. PMID: 38805752 Free PMC article.
References
-
- Rief M., Gautel M., Oesterhelt F. et al. (1997) Reversible unfolding of individual titin immunoglobulin domains by AFM. Science, 276, 1109–1112. - PubMed
-
- Bhasin N., Carl P., Harper S. et al. (2004) Chemistry on a single protein, vascular cell adhesion Molecule-1, during forced unfolding. J. Biol. Chem., 279, 45865–45874. - PubMed
-
- Gelles J. and Landick R. (1998) RNA polymerase as a molecular motor. Cell, 93, 13–16. - PubMed

