T Cells Spatially Regulate B Cell Receptor Signaling in Lymphomas through H3K9me3 Modifications
- PMID: 38837879
- PMCID: PMC11617604
- DOI: 10.1002/adhm.202401192
T Cells Spatially Regulate B Cell Receptor Signaling in Lymphomas through H3K9me3 Modifications
Abstract
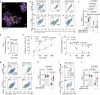
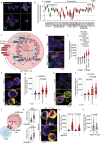
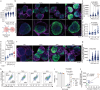
Activated B cell-like diffuse large B-cell lymphoma (ABC-DLBCL) is a subtype associated with poor survival outcomes. Despite identifying therapeutic targets through molecular characterization, targeted therapies have limited success. New strategies using immune-competent tissue models are needed to understand how DLBCL cells evade treatment. Here, synthetic hydrogel-based lymphoma organoids are used to demonstrate how signals in the lymphoid tumor microenvironment (Ly-TME) can alter B cell receptor (BCR) signaling and specific histone modifications, tri-methylation of histone 3 at lysine 9 (H3K9me3), dampening the effects of BCR pathway inhibition. Using imaging modalities, T cells increase DNA methyltransferase 3A expression and cytoskeleton formation in proximal ABC-DLBCL cells, regulated by H3K9me3. Expansion microscopy on lymphoma organoids reveals T cells increase the size and quantity of segregated H3K9me3 clusters in ABC-DLBCL cells. Findings suggest the re-organization of higher-order chromatin structures that may contribute to evasion or resistance to therapy via the emergence of novel transcriptional states. Treating ABC-DLBCL cells with a G9α histone methyltransferase inhibitor reverses T cell-mediated modulation of H3K9me3 and overcomes T cell-mediated attenuation of treatment response to BCR pathway inhibition. This study emphasizes the Ly-TME's role in altering DLBCL fate and suggests targeting aberrant signaling and microenvironmental cross-talk that can benefit high-risk patients.
Keywords: B cell; T cell; immunoengineering; lymphoid; organoids; tumor.
© 2024 The Author(s). Advanced Healthcare Materials published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Sehn L. H., Gascoyne R. D., Blood 2015, 125, 22. - PubMed
-
- Schmitz R., Wright G. W., Huang D. W., Johnson C. A., Phelan J. D., Wang J. Q., Roulland S., Kasbekar M., Young R. M., Shaffer A. L., Hodson D. J., Xiao W., Yu X., Yang Y., Zhao H., Xu W., Liu X., Zhou B., Du W., Chan W. C., Jaffe E. S., Gascoyne R. D., Connors J. M., Campo E., Lopez‐Guillermo A., Rosenwald A., Ott G., Delabie J., Rimsza L. M., Tay Kuang Wei K., et al., N. Engl. J. Med. 2018, 378, 1396. - PMC - PubMed
-
- Phelan J. D., Young R. M., Webster D. E., Roulland S., Wright G. W., Kasbekar M., Shaffer A. L., Ceribelli M., Wang J. Q., Schmitz R., Nakagawa M., Bachy E., Huang D. W., Ji Y., Chen L., Yang Y., Zhao H., Yu X., Xu W., Palisoc M. M., Valadez R. R., Davies‐Hill T., Wilson W. H., Chan W. C., Jaffe E. S., Gascoyne R. D., Campo E., Rosenwald A., Ott G., Delabie J., et al., Nature 2018, 560, 387. - PMC - PubMed
-
- Burger J. A., Wiestner A., Nat. Rev. Cancer 2018, 18, 148. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

