Concurrent RB1 Loss and BRCA Deficiency Predicts Enhanced Immunologic Response and Long-term Survival in Tubo-ovarian High-grade Serous Carcinoma
- PMID: 38837893
- PMCID: PMC11325151
- DOI: 10.1158/1078-0432.CCR-23-3552
Concurrent RB1 Loss and BRCA Deficiency Predicts Enhanced Immunologic Response and Long-term Survival in Tubo-ovarian High-grade Serous Carcinoma
Abstract
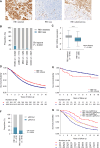
Purpose: The purpose of this study was to evaluate RB1 expression and survival across ovarian carcinoma histotypes and how co-occurrence of BRCA1 or BRCA2 (BRCA) alterations and RB1 loss influences survival in tubo-ovarian high-grade serous carcinoma (HGSC).
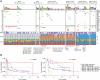
Experimental design: RB1 protein expression was classified by immunohistochemistry in ovarian carcinomas of 7,436 patients from the Ovarian Tumor Tissue Analysis consortium. We examined RB1 expression and germline BRCA status in a subset of 1,134 HGSC, and related genotype to overall survival (OS), tumor-infiltrating CD8+ lymphocytes, and transcriptomic subtypes. Using CRISPR-Cas9, we deleted RB1 in HGSC cells with and without BRCA1 alterations to model co-loss with treatment response. We performed whole-genome and transcriptome data analyses on 126 patients with primary HGSC to characterize tumors with concurrent BRCA deficiency and RB1 loss.
Results: RB1 loss was associated with longer OS in HGSC but with poorer prognosis in endometrioid ovarian carcinoma. Patients with HGSC harboring both RB1 loss and pathogenic germline BRCA variants had superior OS compared with patients with either alteration alone, and their median OS was three times longer than those without pathogenic BRCA variants and retained RB1 expression (9.3 vs. 3.1 years). Enhanced sensitivity to cisplatin and paclitaxel was seen in BRCA1-altered cells with RB1 knockout. Combined RB1 loss and BRCA deficiency correlated with transcriptional markers of enhanced IFN response, cell-cycle deregulation, and reduced epithelial-mesenchymal transition. CD8+ lymphocytes were most prevalent in BRCA-deficient HGSC with co-loss of RB1.
Conclusions: Co-occurrence of RB1 loss and BRCA deficiency was associated with exceptionally long survival in patients with HGSC, potentially due to better treatment response and immune stimulation.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
F.A.M. Saner reports research grants from Swiss National Foundation (Early Postdoc Mobility Fellowship P2BEP3-172246), Swiss Cancer League (grant BIL KFS-3942-08-2016), Prof. Max Cloëtta Foundation grant, and the Foundation for Clinical-Experimental Cancer Research Bern during the conduct of the study. T.A. Zwimpfer reports grants from Swiss National Foundation and Gottfried & Julia Bangerter–Rhyner–Stiftung during the conduct of the study. N.S. Meagher reports grants from NanoString Technologies outside the submitted work. S. Fereday reports grants from AstraZeneca outside the submitted work. N. Traficante reports grants from AstraZeneca Pty. Ltd. outside the submitted work. K. Alsop reports grants from AstraZeneca outside the submitted work. E.L. Christie reports grants from AstraZeneca and personal fees from GSK outside the submitted work. J. Boros reports grants from Cancer Institute New South Wales and National Health and Medical Research Council during the conduct of the study. A. Brooks-Wilson reports grants from Canadian Institutes of Health Research during the conduct of the study. K.L. Cushing-Haugen reports grants from NCI (R01 CA168758) during the conduct of the study. A. Gentry-Maharaj reports other support from intelligent Lab on Fiber (iLoF), RNA Guardian, Micronoma, and Mercy BioAnalytics outside the submitted work, as well as being a member of CRUK ACED Gynaecological Cancer Working Group and CRUK ACED Co-Director Research Domain Trials. A. Hartmann reports personal fees from Eisai and grants from Owkin outside the submitted work. M.E. Jones reports grants from Breast Cancer Now during the conduct of the study. C.J. Kennedy reports grants from National Health and Medical Research Council Enabling Grants ID 310670 and 628903 and Cancer Institute New South Wales Grants ID 12/RIG/1-17 and 15/RIG/1-16 during the conduct of the study. M.J. Schoemaker reports grants from Breast Cancer Now (charity) during the conduct of the study, as well as other support from IQVIA outside the submitted work. M.E. Sherman reports grants from NIH during the conduct of the study and nonfinancial support from Exact Sciences outside the submitted work. M.S. Anglesio reports grants from Michael Smith Health Research BC during the conduct of the study. J.D. Brenton reports being an inventor of patent “Enhanced Detection of Target DNA by Fragment Size Analysis” (WO/2020/094775), patent “TAm-Seq v2 Method for ctDNA Estimation” (WO 2016/009224A1), and patent “Methods for Predicting Treatment Response in Cancers” (patent application no. 1818159.5); being a founder and director of Tailor Bio Ltd.; holding shares in Tailor Bio Ltd; and receiving honoraria and personal payments from AstraZeneca, GSK, and Clovis Oncology. J.A. Doherty reports grants from NCI during the conduct of the study. P.A. Fasching reports grants and personal fees from Novartis and grants from BioNTech and Guardant Health outside the submitted work. R.T. Fortner reports grants from German Federal Ministry of Education and Research, Programme of Clinical Biomedical Research, during the conduct of the study. B.Y. Karlan reports other support from GOG Foundation and Sandy Rollman Foundation outside the submitted work. U. Menon reports patent no. EP10178345.4 for Breast Cancer Diagnostics issued. F. Modugno reports grants from NCI and Department of Defense during the conduct of the study. P.D.P. Pharoah reports grants from US Department of Defense and Cancer Research UK during the conduct of the study. A.J. Swerdlow reports grants from Breast Cancer Now, charity and grants from Ovarian Cancer Action, and charity during the conduct of the study, as well as reports that A.J. Swerdlow's late mother held shares in GSK and Haleon. A. DeFazio reports grants from National Health and Medical Research Council of Australia, Cancer Institute NSW, and US Army Medical Research and Materiel Command during the conduct of the study, as well as grants and nonfinancial support from AstraZeneca and Illumina outside the submitted work. M. Köbel reports personal fees from Helixbiopharma outside the submitted work. D.D.L. Bowtell reports grants from Australian National Health and Medical Research Council and US Department of Defense during the conduct of the study, as well as grants from Genentech Roche and AstraZeneca and personal fees from Exo Therapeutics outside the submitted work. D.W. Garsed reports grants from National Health and Medical Research Council of Australia, US Army Medical Research and Materiel Command Ovarian Cancer Research Program, and Victorian Cancer Agency during the conduct of the study. No disclosures were reported by the other authors.
Figures


Update of
-
Concurrent RB1 loss and BRCA-deficiency predicts enhanced immunological response and long-term survival in tubo-ovarian high-grade serous carcinoma.medRxiv [Preprint]. 2023 Nov 10:2023.11.09.23298321. doi: 10.1101/2023.11.09.23298321. medRxiv. 2023. Update in: Clin Cancer Res. 2024 Aug 15;30(16):3481-3498. doi: 10.1158/1078-0432.CCR-23-3552. PMID: 37986741 Free PMC article. Updated. Preprint.
References
-
- Patch AM, Christie EL, Etemadmoghadam D, Garsed DW, George J, Fereday S, et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature 2015;521:489–94. - PubMed
-
- Burdett NL, Willis MO, Alsop K, Hunt AL, Pandey A, Hamilton PT, et al. Multiomic analysis of homologous recombination-deficient end-stage high-grade serous ovarian cancer. Nat Genet 2023;55:437–50. - PubMed
MeSH terms
Substances
Grants and funding
- P2BEP3-172246/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (SNF)
- Swedish Cancer Foundation
- MCRF22018/Victorian Cancer Agency (VCA)
- Foundation for Clinical-Experimental Cancer Research
- R01CA172404/National Cancer Institute (NCI)
- UL1 TR000124/TR/NCATS NIH HHS/United States
- 742432/HORIZON EUROPE European Research Council (ERC)
- 1186505/National Health and Medical Research Council (NHMRC)
- 2009840/National Health and Medical Research Council (NHMRC)
- 18274/Michael Smith Health Research BC (MSFHR)
- P30 CA015083/CA/NCI NIH HHS/United States
- W81XWH-16-2-0010/U.S. Army Medical Research and Development Command (MRDC)
- UL1 TR001881/TR/NCATS NIH HHS/United States
- BIL KFS-3942-08-2016/Krebsliga Schweiz (Swiss Cancer League)
- MCRF21004/Victorian Cancer Agency (VCA)
- APP1111032/National Health and Medical Research Council (NHMRC)
- R01 CA172404/CA/NCI NIH HHS/United States
- 1092856/National Health and Medical Research Council (NHMRC)
- Janet D. Cottrelle Foundation (JDCF)
- W81XWH-21-1-0401/U.S. Army Medical Research and Development Command (MRDC)
- R01 CA248288/CA/NCI NIH HHS/United States
- P30 CA071789/CA/NCI NIH HHS/United States
- P50 CA136393/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

