CheRRI-Accurate classification of the biological relevance of putative RNA-RNA interaction sites
- PMID: 38837942
- PMCID: PMC11152173
- DOI: 10.1093/gigascience/giae022
CheRRI-Accurate classification of the biological relevance of putative RNA-RNA interaction sites
Abstract
Background: RNA-RNA interactions are key to a wide range of cellular functions. The detection of potential interactions helps to understand the underlying processes. However, potential interactions identified via in silico or experimental high-throughput methods can lack precision because of a high false-positive rate.
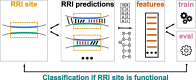
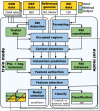
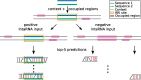
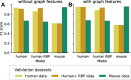
Results: We present CheRRI, the first tool to evaluate the biological relevance of putative RNA-RNA interaction sites. CheRRI filters candidates via a machine learning-based model trained on experimental RNA-RNA interactome data. Its unique setup combines interactome data and an established thermodynamic prediction tool to integrate experimental data with state-of-the-art computational models. Applying these data to an automated machine learning approach provides the opportunity to not only filter data for potential false positives but also tailor the underlying interaction site model to specific needs.
Conclusions: CheRRI is a stand-alone postprocessing tool to filter either predicted or experimentally identified potential RNA-RNA interactions on a genomic level to enhance the quality of interaction candidates. It is easy to install (via conda, pip packages), use (via Galaxy), and integrate into existing RNA-RNA interaction pipelines.
Keywords: RNA–RNA interactome; classification; direct duplex detection; false positives; functional RRI.
© The Author(s) 2024. Published by Oxford University Press GigaScience.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

