ADH1B, the adipocyte-enriched alcohol dehydrogenase, plays an essential, cell-autonomous role in human adipogenesis
- PMID: 38838011
- PMCID: PMC11181076
- DOI: 10.1073/pnas.2319301121
ADH1B, the adipocyte-enriched alcohol dehydrogenase, plays an essential, cell-autonomous role in human adipogenesis
Abstract
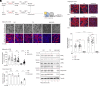
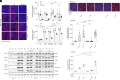
Alcohol dehydrogenase 1B (ADH1B) is a primate-specific enzyme which, uniquely among the ADH class 1 family, is highly expressed both in adipose tissue and liver. Its expression in adipose tissue is reduced in obesity and increased by insulin stimulation. Interference with ADH1B expression has also been reported to impair adipocyte function. To better understand the role of ADH1B in adipocytes, we used CRISPR/Cas9 to delete ADH1B in human adipose stem cells (ASC). Cells lacking ADH1B failed to differentiate into mature adipocytes manifested by minimal triglyceride accumulation and a marked reduction in expression of established adipocyte markers. As ADH1B is capable of converting retinol to retinoic acid (RA), we conducted rescue experiments. Incubation of ADH1B-deficient preadipocytes with 9-cis-RA, but not with all-transretinol, significantly rescued their ability to accumulate lipids and express markers of adipocyte differentiation. A homozygous missense variant in ADH1B (p.Arg313Cys) was found in a patient with congenital lipodystrophy of unknown cause. This variant significantly impaired the protein's dimerization, enzymatic activity, and its ability to rescue differentiation in ADH1B-deficient ASC. The allele frequency of this variant in the Middle Eastern population suggests that it is unlikely to be a fully penetrant cause of severe lipodystrophy. In conclusion, ADH1B appears to play an unexpected, crucial and cell-autonomous role in human adipocyte differentiation by serving as a necessary source of endogenous retinoic acid.
Keywords: 9-cis retinoic acid; ADH1B; adipocyte differentiation; alcohol dehydrogenase 1B; human adipose stem cells.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures


References
-
- Smith M., Hopkinson D. A., Harris H., Alcohol dehydrogenase isozymes in adult human stomach and liver: Evidence for activity of the ADH 3 locus. Ann. Hum. Genet. 35, 243–253 (1972). - PubMed
-
- McEvily A. J., Holmquist B., Auld D. S., Vallee B. L., 3 beta-Hydroxy-5 beta-steroid dehydrogenase activity of human liver alcohol dehydrogenase is specific to gamma-subunits. Biochemistry 27, 4284–4288 (1988). - PubMed
-
- Stone C. L., Li T. K., Bosron W. F., Stereospecific oxidation of secondary alcohols by human alcohol dehydrogenases. J. Biol. Chem. 264, 11112–11116 (1989). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

