Single-Cell Spatial Transcriptomics Unveils Platelet-Fueled Cycling Macrophages for Kidney Fibrosis
- PMID: 38838052
- PMCID: PMC11304276
- DOI: 10.1002/advs.202308505
Single-Cell Spatial Transcriptomics Unveils Platelet-Fueled Cycling Macrophages for Kidney Fibrosis
Abstract
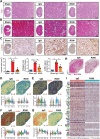
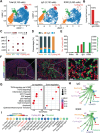
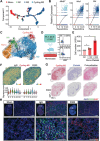
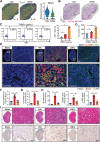
With the increasing incidence of kidney diseases, there is an urgent need to develop therapeutic strategies to combat post-injury fibrosis. Immune cells, including platelets, play a pivotal role in this repair process, primarily through their released cytokines. However, the specific role of platelets in kidney injury and subsequent repair remains underexplored. Here, the detrimental role of platelets in renal recovery following ischemia/reperfusion injury and its contribution to acute kidney injury to chronic kidney disease transition is aimed to investigated. In this study, it is shown that depleting platelets accelerates injury resolution and significantly reduces fibrosis. Employing advanced single-cell and spatial transcriptomic techniques, macrophages as the primary mediators modulated by platelet signals is identified. A novel subset of macrophages, termed "cycling M2", which exhibit an M2 phenotype combined with enhanced proliferative activity is uncovered. This subset emerges in the injured kidney during the resolution phase and is modulated by platelet-derived thrombospondin 1 (THBS1) signaling, acquiring profibrotic characteristics. Conversely, targeted inhibition of THBS1 markedly downregulates the cycling M2 macrophage, thereby mitigating fibrotic progression. Overall, this findings highlight the adverse role of platelet THBS1-boosted cycling M2 macrophages in renal injury repair and suggest platelet THBS1 as a promising therapeutic target for alleviating inflammation and kidney fibrosis.
Keywords: kidney fibrosis; macrophage proliferation; platelet; thrombospondin 1.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Kellum J. A., Romagnani P., Ashuntantang G., Ronco C., Zarbock A., Anders H. J., Nat. Rev. Dis. Primers. 2020, 7, 52. - PubMed
-
- Hoste E. A. J., Kellum J. A., Selby N. M., Zarbock A., Palevsky P. M., Bagshaw S. M., Goldstein S. L., Cerdá J., Chawla L. S., Nat. Rev. Nephrol. 2018, 14, 607. - PubMed
-
- Meng X. M., Nikolic‐Paterson D. J., Lan H. Y., Nat. Rev. Nephrol. 2016, 12, 325. - PubMed
MeSH terms
Grants and funding
- 82270713/National Natural Science Foundation of China
- SZS2022018/Key Laboratory Foundation of Structural Deformities in Children of Suzhou
- GSWS2022058/Gusu Health Talent Project of Suzhou
- GSWS2020049/Gusu Health Talent Project of Suzhou
- SKY2023059/Suzhou Science and Technology Development Plan Project
LinkOut - more resources
Full Text Sources
Miscellaneous
