Virological characteristics of a SARS-CoV-2-related bat coronavirus, BANAL-20-236
- PMID: 38838469
- PMCID: PMC11215962
- DOI: 10.1016/j.ebiom.2024.105181
Virological characteristics of a SARS-CoV-2-related bat coronavirus, BANAL-20-236
Abstract
Background: Although several SARS-CoV-2-related coronaviruses (SC2r-CoVs) were discovered in bats and pangolins, the differences in virological characteristics between SARS-CoV-2 and SC2r-CoVs remain poorly understood. Recently, BANAL-20-236 (B236) was isolated from a rectal swab of Malayan horseshoe bat and was found to lack a furin cleavage site (FCS) in the spike (S) protein. The comparison of its virological characteristics with FCS-deleted SARS-CoV-2 (SC2ΔFCS) has not been conducted yet.
Methods: We prepared human induced pluripotent stem cell (iPSC)-derived airway and lung epithelial cells and colon organoids as human organ-relevant models. B236, SARS-CoV-2, and artificially generated SC2ΔFCS were used for viral experiments. To investigate the pathogenicity of B236 in vivo, we conducted intranasal infection experiments in hamsters.
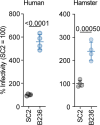
Findings: In human iPSC-derived airway epithelial cells, the growth of B236 was significantly lower than that of the SC2ΔFCS. A fusion assay showed that the B236 and SC2ΔFCS S proteins were less fusogenic than the SARS-CoV-2 S protein. The infection experiment in hamsters showed that B236 was less pathogenic than SARS-CoV-2 and even SC2ΔFCS. Interestingly, in human colon organoids, the growth of B236 was significantly greater than that of SARS-CoV-2.
Interpretation: Compared to SARS-CoV-2, we demonstrated that B236 exhibited a tropism toward intestinal cells rather than respiratory cells. Our results are consistent with a previous report showing that B236 is enterotropic in macaques. Altogether, our report strengthens the assumption that SC2r-CoVs in horseshoe bats replicate primarily in the intestinal tissues rather than respiratory tissues.
Funding: This study was supported in part by AMED ASPIRE (JP23jf0126002, to Keita Matsuno, Kazuo Takayama, and Kei Sato); AMED SCARDA Japan Initiative for World-leading Vaccine Research and Development Centers "UTOPIA" (JP223fa627001, to Kei Sato), AMED SCARDA Program on R&D of new generation vaccine including new modality application (JP223fa727002, to Kei Sato); AMED SCARDA Hokkaido University Institute for Vaccine Research and Development (HU-IVReD) (JP223fa627005h0001, to Takasuke Fukuhara, and Keita Matsuno); AMED Research Program on Emerging and Re-emerging Infectious Diseases (JP21fk0108574, to Hesham Nasser; JP21fk0108493, to Takasuke Fukuhara; JP22fk0108617 to Takasuke Fukuhara; JP22fk0108146, to Kei Sato; JP21fk0108494 to G2P-Japan Consortium, Keita Matsuno, Shinya Tanaka, Terumasa Ikeda, Takasuke Fukuhara, and Kei Sato; JP21fk0108425, to Kazuo Takayama and Kei Sato; JP21fk0108432, to Kazuo Takayama, Takasuke Fukuhara and Kei Sato; JP22fk0108534, Terumasa Ikeda, and Kei Sato; JP22fk0108511, to Yuki Yamamoto, Terumasa Ikeda, Keita Matsuno, Shinya Tanaka, Kazuo Takayama, Takasuke Fukuhara, and Kei Sato; JP22fk0108506, to Kazuo Takayama and Kei Sato); AMED Research Program on HIV/AIDS (JP22fk0410055, to Terumasa Ikeda; and JP22fk0410039, to Kei Sato); AMED Japan Program for Infectious Diseases Research and Infrastructure (JP22wm0125008 to Keita Matsuno); AMED CREST (JP21gm1610005, to Kazuo Takayama; JP22gm1610008, to Takasuke Fukuhara; JST PRESTO (JPMJPR22R1, to Jumpei Ito); JST CREST (JPMJCR20H4, to Kei Sato); JSPS KAKENHI Fund for the Promotion of Joint International Research (International Leading Research) (JP23K20041, to G2P-Japan Consortium, Keita Matsuno, Takasuke Fukuhara and Kei Sato); JST SPRING (JPMJSP2108 to Shigeru Fujita); JSPS KAKENHI Grant-in-Aid for Scientific Research C (22K07103, to Terumasa Ikeda); JSPS KAKENHI Grant-in-Aid for Scientific Research B (21H02736, to Takasuke Fukuhara); JSPS KAKENHI Grant-in-Aid for Early-Career Scientists (22K16375, to Hesham Nasser; 20K15767, to Jumpei Ito); JSPS Core-to-Core Program (A. Advanced Research Networks) (JPJSCCA20190008, to Kei Sato); JSPS Research Fellow DC2 (22J11578, to Keiya Uriu); JSPS Research Fellow DC1 (23KJ0710, to Yusuke Kosugi); JSPS Leading Initiative for Excellent Young Researchers (LEADER) (to Terumasa Ikeda); World-leading Innovative and Smart Education (WISE) Program 1801 from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) (to Naganori Nao); Ministry of Health, Labour and Welfare (MHLW) under grant 23HA2010 (to Naganori Nao and Keita Matsuno); The Cooperative Research Program (Joint Usage/Research Center program) of Institute for Life and Medical Sciences, Kyoto University (to Kei Sato); International Joint Research Project of the Institute of Medical Science, the University of Tokyo (to Terumasa Ikeda and Takasuke Fukuhara); The Tokyo Biochemical Research Foundation (to Kei Sato); Takeda Science Foundation (to Terumasa Ikeda and Takasuke Fukuhara); Mochida Memorial Foundation for Medical and Pharmaceutical Research (to Terumasa Ikeda); The Naito Foundation (to Terumasa Ikeda); Hokuto Foundation for Bioscience (to Tomokazu Tamura); Hirose Foundation (to Tomokazu Tamura); and Mitsubishi Foundation (to Kei Sato).
Keywords: BANAL-20-236; Bat coronavirus; SARS-CoV-2; Spillover.
Copyright © 2024 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests Yuki Yamamoto and Tetsuharu Nagamoto are founders and shareholders of HiLung, Inc. Yuki Yamamoto is a co-inventor of patents (PCT/JP2016/057254; "Method for inducing differentiation of alveolar epithelial cells", PCT/JP2016/059786, "Method of producing airway epithelial cells"). Jumpei Ito has consulting fees and honoraria for lectures from Takeda Pharmaceutical Co. Ltd. Kei Sato has consulting fees from Moderna Japan Co., Ltd. and Takeda Pharmaceutical Co. Ltd. and honoraria for lectures from Gilead Sciences, Inc., Moderna Japan Co., Ltd., and Shionogi & Co., Ltd. The other authors declare that no competing interests exist.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

