Epithelial hypoxia maintains colonization resistance against Candida albicans
- PMID: 38838675
- PMCID: PMC11239274
- DOI: 10.1016/j.chom.2024.05.008
Epithelial hypoxia maintains colonization resistance against Candida albicans
Abstract
Antibiotic treatment promotes the outgrowth of intestinal Candida albicans, but the mechanisms driving this fungal bloom remain incompletely understood. We identify oxygen as a resource required for post-antibiotic C. albicans expansion. C. albicans depleted simple sugars in the ceca of gnotobiotic mice but required oxygen to grow on these resources in vitro, pointing to anaerobiosis as a potential factor limiting growth in the gut. Clostridia species limit oxygen availability in the large intestine by producing butyrate, which activates peroxisome proliferator-activated receptor gamma (PPAR-γ) signaling to maintain epithelial hypoxia. Streptomycin treatment depleted Clostridia-derived butyrate to increase epithelial oxygenation, but the PPAR-γ agonist 5-aminosalicylic acid (5-ASA) functionally replaced Clostridia species to restore epithelial hypoxia and colonization resistance against C. albicans. Additionally, probiotic Escherichia coli required oxygen respiration to prevent a post-antibiotic bloom of C. albicans, further supporting the role of oxygen in colonization resistance. We conclude that limited access to oxygen maintains colonization resistance against C. albicans.
Keywords: Candida albicans; Clostridia; Escherichia coli; colonization resistance; epithelial hypoxia.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests G.R.T. is consulting for Astellas, Cidara, F2G, Immy, Mayne, Melinta, Mundipharma, Scynexis, and Pfizer on projects that are not related to this publication. G.R.T. receives research support from Astellas, Cidara, F2G, Mayne, Melinta, Merck, Mundipharma, Scynexis, and Pfizer for projects that are not related to this publication.
Figures
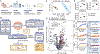
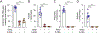

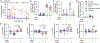


References
MeSH terms
Substances
Grants and funding
- R56 AI112949/AI/NIAID NIH HHS/United States
- TL1 TR000133/TR/NCATS NIH HHS/United States
- R01 AI044170/AI/NIAID NIH HHS/United States
- R21 AI146432/AI/NIAID NIH HHS/United States
- R01 AI096528/AI/NIAID NIH HHS/United States
- R21 AI153069/AI/NIAID NIH HHS/United States
- UL1 TR000002/TR/NCATS NIH HHS/United States
- UL1 TR001860/TR/NCATS NIH HHS/United States
- R01 AI112949/AI/NIAID NIH HHS/United States
- R01 DK138912/DK/NIDDK NIH HHS/United States
- F32 AI161850/AI/NIAID NIH HHS/United States
- R01 AI112445/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources

