Integrating system biology and intratumor gene therapy by trans-complementing the appropriate co-stimulatory molecule as payload in oncolytic herpes virus
- PMID: 38839891
- PMCID: PMC11405262
- DOI: 10.1038/s41417-024-00790-8
Integrating system biology and intratumor gene therapy by trans-complementing the appropriate co-stimulatory molecule as payload in oncolytic herpes virus
Abstract
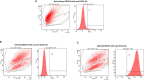
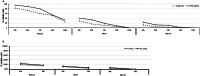
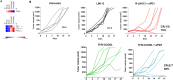
Systems biology has been applied at the multi-scale level within the cancer field, improving cancer prevention, diagnosis and enabling precision medicine approaches. While systems biology can expand the knowledge and skills for oncological treatment, it also represents a challenging expedition due to cancer complexity, heterogeneity and diversity not only between different cancer indications, but also in its evolution process through space and time. Here, by characterizing the transcriptional perturbations of the tumor microenvironment induced by oncolytic, we aimed to rationally design a novel armed oncolytic herpes virus. We found that intratumor oncovirotherapy with HSV-1 induces T-cell activation signatures and transcriptionally activates several costimulatory molecules. We identified differentially expressed costimulatory receptors and binding partners, where inducible co-stimulators (ICOS) resulted in the potentially most beneficial targeted therapy. Through an ex-vivo transcriptomic analysis, we explored the potential of arming an oncolytic virus as a combination therapy strategy; in particular, we engineered a targeted herpes virus encoding ICOSL (THV_ICOSL), which resulted in a significant improvement in tumor size control compared to unarmed parental virus. Also, combination with a PD-1 inhibitor enhanced antitumor efficacy as predictable by upregulation of PD-1 and ligands pair (PD-L1/PD-L2) upon oncolytic virus injection. Generation of the human version of this virus encoding hICOSL orthologue effectively and specifically activated human T cells by triggering the ICOS pathway. Our data support the data-driven generation of armed oncolytic viruses as combination immunotherapeutic with checkpoint inhibitors.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Development of a new fusion-enhanced oncolytic immunotherapy platform based on herpes simplex virus type 1.J Immunother Cancer. 2019 Aug 10;7(1):214. doi: 10.1186/s40425-019-0682-1. J Immunother Cancer. 2019. PMID: 31399043 Free PMC article.
-
Oncolytic virus therapy using genetically engineered herpes simplex viruses.Front Biosci. 2008 Jan 1;13:2060-4. doi: 10.2741/2823. Front Biosci. 2008. PMID: 17981691 Review.
-
Generation of a Retargeted Oncolytic Herpes Virus Encoding Adenosine Deaminase for Tumor Adenosine Clearance.Int J Mol Sci. 2021 Dec 16;22(24):13521. doi: 10.3390/ijms222413521. Int J Mol Sci. 2021. PMID: 34948316 Free PMC article.
-
Intratumoral Delivery of a PD-1-Blocking scFv Encoded in Oncolytic HSV-1 Promotes Antitumor Immunity and Synergizes with TIGIT Blockade.Cancer Immunol Res. 2020 May;8(5):632-647. doi: 10.1158/2326-6066.CIR-19-0628. Epub 2020 Mar 3. Cancer Immunol Res. 2020. PMID: 32127389
-
Advance in herpes simplex viruses for cancer therapy.Sci China Life Sci. 2013 Apr;56(4):298-305. doi: 10.1007/s11427-013-4466-4. Epub 2013 Apr 7. Sci China Life Sci. 2013. PMID: 23564184 Review.
Cited by
-
We ARE boosting translation: AU-rich elements for enhanced therapeutic mRNA translation.Mol Ther Nucleic Acids. 2025 Apr 14;36(2):102531. doi: 10.1016/j.omtn.2025.102531. eCollection 2025 Jun 10. Mol Ther Nucleic Acids. 2025. PMID: 40276699 Free PMC article. No abstract available.
References
-
- Sasso E, Paciello R, D’Auria F, Riccio G, Froechlich G, Cortese R, et al. One-step recovery of scFv clones from high-throughput sequencing-based screening of phage display libraries challenged to cells expressing native claudin-1. Biomed Res Int. 2015;2015:703213. 10.1155/2015/703213 - DOI - PMC - PubMed
-
- Zambrano N, Froechlich G, Lazarevic D, Passariello M, Nicosia A, De Lorenzo C, et al. High-throughput monoclonal antibody discovery from phage libraries: challenging the current preclinical pipeline to keep the pace with the increasing mAb demand. Cancers. 2022;14:1325. 10.3390/cancers14051325 - DOI - PMC - PubMed
-
- Esposito MV, Minopoli G, Esposito L, D’Argenio V, Di Maggio F, Sasso E, et al. A functional analysis of the unclassified Pro2767Ser BRCA2 variant reveals its potential pathogenicity that acts by hampering DNA binding and homology-mediated DNA repair. Cancers. 2019;11:1454. 10.3390/cancers11101454 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

