Prevalence of coronaviruses in European bison (Bison bonasus) in Poland
- PMID: 38839918
- PMCID: PMC11153543
- DOI: 10.1038/s41598-024-63717-1
Prevalence of coronaviruses in European bison (Bison bonasus) in Poland
Abstract
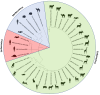
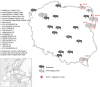
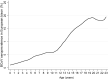
Coronaviruses have been confirmed to infect a variety of species, but only one case of associated winter dysentery of European bison has been described. The study aimed to analyze the prevalence, and define the impact on the species conservation, the source of coronavirus infection, and the role of the European bison in the transmission of the pathogen in Poland. Molecular and serological screening was performed on 409 European bison from 6 free-ranging and 14 captive herds over the period of 6 years (2017-2023). Presence of coronavirus was confirmed in one nasal swab by pancoronavirus RT-PCR and in 3 nasal swab samples by bovine coronavirus (BCoV) specific real time RT-PCR. The detected virus showed high (> 98%) homology in both RdRp and Spike genes to BCoV strains characterised recently in Polish cattle and strains isolated from wild cervids in Italy. Antibodies specific to BCoV were found in 6.4% of tested samples, all originating from free-ranging animals. Seroprevalence was higher in adult animals over 5 years of age (p = 0.0015) and in females (p = 0.09). Our results suggest that European bison play only a limited role as reservoirs of bovine-like coronaviruses. Although the most probable source of infections in the European bison population in Poland is cattle, other wild ruminants could also be involved. In addition, the zoonotic potential of bovine coronaviruses is quite low.
Keywords: BCoV; Coronaviruses; European bison; PCR; Sanger sequencing; Seroprevalence.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
A serological survey of pathogens associated with the respiratory and digestive system in the Polish European bison (Bison bonasus) population in 2017-2022.BMC Vet Res. 2023 Jun 1;19(1):74. doi: 10.1186/s12917-023-03627-y. BMC Vet Res. 2023. PMID: 37264393 Free PMC article.
-
Presence of anti-SARS-CoV-2 antibodies in European bison (Bison bonasus) in Poland, 2019-2023.BMC Vet Res. 2025 Feb 28;21(1):120. doi: 10.1186/s12917-025-04593-3. BMC Vet Res. 2025. PMID: 40022124 Free PMC article.
-
Biologic, antigenic, and full-length genomic characterization of a bovine-like coronavirus isolated from a giraffe.J Virol. 2007 May;81(10):4981-90. doi: 10.1128/JVI.02361-06. Epub 2007 Mar 7. J Virol. 2007. PMID: 17344285 Free PMC article.
-
Advances in Laboratory Diagnosis of Coronavirus Infections in Cattle.Pathogens. 2024 Jun 21;13(7):524. doi: 10.3390/pathogens13070524. Pathogens. 2024. PMID: 39057751 Free PMC article. Review.
-
Bovine respiratory coronavirus.Vet Clin North Am Food Anim Pract. 2010 Jul;26(2):349-64. doi: 10.1016/j.cvfa.2010.04.005. Vet Clin North Am Food Anim Pract. 2010. PMID: 20619189 Free PMC article. Review.
Cited by
-
Morphological patterns of the European bison (Bison bonasus) skull.Sci Rep. 2025 Jan 9;15(1):1418. doi: 10.1038/s41598-025-85654-3. Sci Rep. 2025. PMID: 39789289 Free PMC article.
-
Machine learning tools used for mapping some immunogenic epitopes within the major structural proteins of the bovine coronavirus (BCoV) and for the in silico design of the multiepitope-based vaccines.Front Vet Sci. 2024 Oct 2;11:1468890. doi: 10.3389/fvets.2024.1468890. eCollection 2024. Front Vet Sci. 2024. PMID: 39415947 Free PMC article.
References
-
- Krasińska M, Krasiński ZA, Tokarska M. Żubr: Monografia Przyrodnicza. Chyra.pl; 2017.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

