Epigenetic inheritance of diet-induced and sperm-borne mitochondrial RNAs
- PMID: 38839949
- PMCID: PMC11186758
- DOI: 10.1038/s41586-024-07472-3
Epigenetic inheritance of diet-induced and sperm-borne mitochondrial RNAs
Abstract
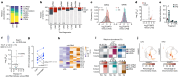
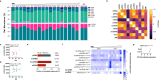
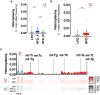
Spermatozoa harbour a complex and environment-sensitive pool of small non-coding RNAs (sncRNAs)1, which influences offspring development and adult phenotypes1-7. Whether spermatozoa in the epididymis are directly susceptible to environmental cues is not fully understood8. Here we used two distinct paradigms of preconception acute high-fat diet to dissect epididymal versus testicular contributions to the sperm sncRNA pool and offspring health. We show that epididymal spermatozoa, but not developing germ cells, are sensitive to the environment and identify mitochondrial tRNAs (mt-tRNAs) and their fragments (mt-tsRNAs) as sperm-borne factors. In humans, mt-tsRNAs in spermatozoa correlate with body mass index, and paternal overweight at conception doubles offspring obesity risk and compromises metabolic health. Sperm sncRNA sequencing of mice mutant for genes involved in mitochondrial function, and metabolic phenotyping of their wild-type offspring, suggest that the upregulation of mt-tsRNAs is downstream of mitochondrial dysfunction. Single-embryo transcriptomics of genetically hybrid two-cell embryos demonstrated sperm-to-oocyte transfer of mt-tRNAs at fertilization and suggested their involvement in the control of early-embryo transcription. Our study supports the importance of paternal health at conception for offspring metabolism, shows that mt-tRNAs are diet-induced and sperm-borne and demonstrates, in a physiological setting, father-to-offspring transfer of sperm mitochondrial RNAs at fertilization.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures









References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

