Senescent glia link mitochondrial dysfunction and lipid accumulation
- PMID: 38839958
- PMCID: PMC11168935
- DOI: 10.1038/s41586-024-07516-8
Senescent glia link mitochondrial dysfunction and lipid accumulation
Erratum in
-
Author Correction: Senescent glia link mitochondrial dysfunction and lipid accumulation.Nature. 2024 Aug;632(8026):E8. doi: 10.1038/s41586-024-07904-0. Nature. 2024. PMID: 39112716 Free PMC article. No abstract available.
Abstract
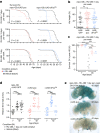
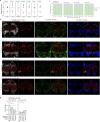
Senescence is a cellular state linked to ageing and age-onset disease across many mammalian species1,2. Acutely, senescent cells promote wound healing3,4 and prevent tumour formation5; but they are also pro-inflammatory, thus chronically exacerbate tissue decline. Whereas senescent cells are active targets for anti-ageing therapy6-11, why these cells form in vivo, how they affect tissue ageing and the effect of their elimination remain unclear12,13. Here we identify naturally occurring senescent glia in ageing Drosophila brains and decipher their origin and influence. Using Activator protein 1 (AP1) activity to screen for senescence14,15, we determine that senescent glia can appear in response to neuronal mitochondrial dysfunction. In turn, senescent glia promote lipid accumulation in non-senescent glia; similar effects are seen in senescent human fibroblasts in culture. Targeting AP1 activity in senescent glia mitigates senescence biomarkers, extends fly lifespan and health span, and prevents lipid accumulation. However, these benefits come at the cost of increased oxidative damage in the brain, and neuronal mitochondrial function remains poor. Altogether, our results map the trajectory of naturally occurring senescent glia in vivo and indicate that these cells link key ageing phenomena: mitochondrial dysfunction and lipid accumulation.
© 2024. The Author(s).
Conflict of interest statement
G.C. is the Director of the Merck-Purdue Center funded by Merck Sharp & Dohme, a subsidiary of Merck, and is a cofounder of Meditati Inc. and BrainGnosis Inc. The remaining authors declare no competing interests.
Figures













References
MeSH terms
Substances
Grants and funding
- RF1 MH128866/MH/NIMH NIH HHS/United States
- P40 OD018537/OD/NIH HHS/United States
- R01 AG071861/AG/NIA NIH HHS/United States
- F31 NS111868/NS/NINDS NIH HHS/United States
- P30 CA023168/CA/NCI NIH HHS/United States
- U18 TR004146/TR/NCATS NIH HHS/United States
- K99 HL147212/HL/NHLBI NIH HHS/United States
- T32 AG000255/AG/NIA NIH HHS/United States
- R01 NS120960/NS/NINDS NIH HHS/United States
- F32 AG066459/AG/NIA NIH HHS/United States
- P01 AG031862/AG/NIA NIH HHS/United States
- R35 NS097275/NS/NINDS NIH HHS/United States
- K99 AG073450/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

