A disease-associated gene desert directs macrophage inflammation through ETS2
- PMID: 38839969
- PMCID: PMC11168933
- DOI: 10.1038/s41586-024-07501-1
A disease-associated gene desert directs macrophage inflammation through ETS2
Abstract
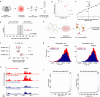
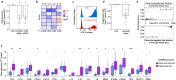
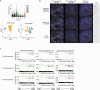
Increasing rates of autoimmune and inflammatory disease present a burgeoning threat to human health1. This is compounded by the limited efficacy of available treatments1 and high failure rates during drug development2, highlighting an urgent need to better understand disease mechanisms. Here we show how functional genomics could address this challenge. By investigating an intergenic haplotype on chr21q22-which has been independently linked to inflammatory bowel disease, ankylosing spondylitis, primary sclerosing cholangitis and Takayasu's arteritis3-6-we identify that the causal gene, ETS2, is a central regulator of human inflammatory macrophages and delineate the shared disease mechanism that amplifies ETS2 expression. Genes regulated by ETS2 were prominently expressed in diseased tissues and more enriched for inflammatory bowel disease GWAS hits than most previously described pathways. Overexpressing ETS2 in resting macrophages reproduced the inflammatory state observed in chr21q22-associated diseases, with upregulation of multiple drug targets, including TNF and IL-23. Using a database of cellular signatures7, we identified drugs that might modulate this pathway and validated the potent anti-inflammatory activity of one class of small molecules in vitro and ex vivo. Together, this illustrates the power of functional genomics, applied directly in primary human cells, to identify immune-mediated disease mechanisms and potential therapeutic opportunities.
© 2024. The Author(s).
Conflict of interest statement
C.T.S., C.B. and J.C.L. are listed as co-inventors on a patent application related to this work. C.W. holds a part-time position at GSK. GSK had no role in the design or conduct of this study. F.N.N. and P.D. are employees and shareholders of NanoString Technologies. NanoString had no role in the design or conduct of this study. The other authors declare no competing interests.
Figures












Comment in
-
ETS2 drives IBD.Nat Rev Drug Discov. 2024 Aug;23(8):581. doi: 10.1038/d41573-024-00110-7. Nat Rev Drug Discov. 2024. PMID: 38937615 No abstract available.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

