MFSD1 with its accessory subunit GLMP functions as a general dipeptide uniporter in lysosomes
- PMID: 38839979
- PMCID: PMC11252000
- DOI: 10.1038/s41556-024-01436-5
MFSD1 with its accessory subunit GLMP functions as a general dipeptide uniporter in lysosomes
Abstract
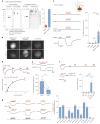
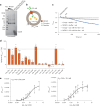
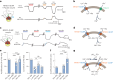
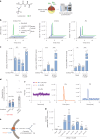
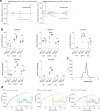
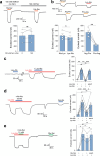
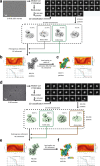
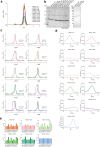
The lysosomal degradation of macromolecules produces diverse small metabolites exported by specific transporters for reuse in biosynthetic pathways. Here we deorphanized the major facilitator superfamily domain containing 1 (MFSD1) protein, which forms a tight complex with the glycosylated lysosomal membrane protein (GLMP) in the lysosomal membrane. Untargeted metabolomics analysis of MFSD1-deficient mouse lysosomes revealed an increase in cationic dipeptides. Purified MFSD1 selectively bound diverse dipeptides, while electrophysiological, isotope tracer and fluorescence-based studies in Xenopus oocytes and proteoliposomes showed that MFSD1-GLMP acts as a uniporter for cationic, neutral and anionic dipeptides. Cryoelectron microscopy structure of the dipeptide-bound MFSD1-GLMP complex in outward-open conformation characterized the heterodimer interface and, in combination with molecular dynamics simulations, provided a structural basis for its selectivity towards diverse dipeptides. Together, our data identify MFSD1 as a general lysosomal dipeptide uniporter, providing an alternative route to recycle lysosomal proteolysis products when lysosomal amino acid exporters are overloaded.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures









References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

