Blastomere size in the human 2-cell embryo predicts the division order that leads to imbalanced lineage contribution to the future body
- PMID: 38841597
- PMCID: PMC11151110
- DOI: 10.17912/micropub.biology.001181
Blastomere size in the human 2-cell embryo predicts the division order that leads to imbalanced lineage contribution to the future body
Abstract
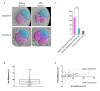
Retrospective tracing of somatic mutations predicted that most cells in the human body could be traced back to a single cell of the 2-cell stage embryo. Accordingly, a recent prospective study of the developmental trajectory of blastomeres in human embryos confirmed that progeny of the first 2-cell stage blastomere to divide generates more epiblast cells (future body). How the 2-cell blastomeres differ is unknown. Here, we show that 2-cell stage blastomeres in human embryos are asymmetric; they differ in size and the bigger blastomere divides first to 4-cell stage. We propose that this asymmetry might originate differences in cell fate.
Copyright: © 2024 by the authors.
Conflict of interest statement
The authors declare that there are no conflicts of interest present.
Figures
References
-
- Bischoff Marcus, Parfitt David-Emlyn, Zernicka-Goetz Magdalena. Formation of the embryonic-abembryonic axis of the mouse blastocyst:relationships between orientation of early cleavage divisions and pattern of symmetric/asymmetric divisions. Development. 2008 Mar 1;135(5):953–962. doi: 10.1242/dev.014316. - DOI - PMC - PubMed
-
- Coorens Tim H. H., Moore Luiza, Robinson Philip S., Sanghvi Rashesh, Christopher Joseph, Hewinson James, Przybilla Moritz J., Lawson Andrew R. J., Spencer Chapman Michael, Cagan Alex, Oliver Thomas R. W., Neville Matthew D. C., Hooks Yvette, Noorani Ayesha, Mitchell Thomas J., Fitzgerald Rebecca C., Campbell Peter J., Martincorena Iñigo, Rahbari Raheleh, Stratton Michael R. Extensive phylogenies of human development inferred from somatic mutations. Nature. 2021 Aug 25;597(7876):387–392. doi: 10.1038/s41586-021-03790-y. - DOI - PubMed
LinkOut - more resources
Full Text Sources
