Host genetics and COVID-19 severity: increasing the accuracy of latest severity scores by Boolean quantum features
- PMID: 38841724
- PMCID: PMC11150643
- DOI: 10.3389/fgene.2024.1362469
Host genetics and COVID-19 severity: increasing the accuracy of latest severity scores by Boolean quantum features
Abstract
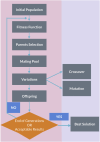
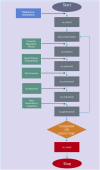
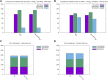
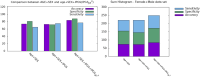
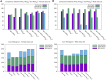
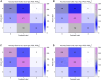
The impact of common and rare variants in COVID-19 host genetics has been widely studied. In particular, in Fallerini et al. (Human genetics, 2022, 141, 147-173), common and rare variants were used to define an interpretable machine learning model for predicting COVID-19 severity. First, variants were converted into sets of Boolean features, depending on the absence or the presence of variants in each gene. An ensemble of LASSO logistic regression models was used to identify the most informative Boolean features with respect to the genetic bases of severity. After that, the Boolean features, selected by these logistic models, were combined into an Integrated PolyGenic Score (IPGS), which offers a very simple description of the contribution of host genetics in COVID-19 severity.. IPGS leads to an accuracy of 55%-60% on different cohorts, and, after a logistic regression with both IPGS and age as inputs, it leads to an accuracy of 75%. The goal of this paper is to improve the previous results, using not only the most informative Boolean features with respect to the genetic bases of severity but also the information on host organs involved in the disease. In this study, we generalize the IPGS adding a statistical weight for each organ, through the transformation of Boolean features into "Boolean quantum features," inspired by quantum mechanics. The organ coefficients were set via the application of the genetic algorithm PyGAD, and, after that, we defined two new integrated polygenic scores ( and ). By applying a logistic regression with both IPGS, ( (or indifferently ) and age as inputs, we reached an accuracy of 84%-86%, thus improving the results previously shown in Fallerini et al. (Human genetics, 2022, 141, 147-173) by a factor of 10%.
Keywords: COVID-19; genetic algorithm; genetic science modeling; host genetics; integrated polygenic score; logistic regression.
Copyright © 2024 Martelloni, Turchi, Fallerini, Degl’Innocenti, Baldassarri, Olmi, Furini, Renieri and GEN-COVID Multicenter study.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Agosto A., Giudici P. (2020). A Poisson autoregressive model to understand covid-19 contagion dynamics. Risks 8, 77. 10.3390/risks8030077 - DOI

