Multiple hybridization events and repeated evolution of homoeologue expression bias in parthenogenetic, polyploid New Zealand stick insects
- PMID: 38842022
- PMCID: PMC12288827
- DOI: 10.1111/mec.17422
Multiple hybridization events and repeated evolution of homoeologue expression bias in parthenogenetic, polyploid New Zealand stick insects
Abstract
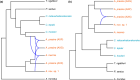
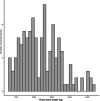
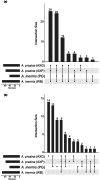
During hybrid speciation, homoeologues combine in a single genome. Homoeologue expression bias (HEB) occurs when one homoeologue has higher gene expression than another. HEB has been well characterized in plants but rarely investigated in animals, especially invertebrates. Consequently, we have little idea as to the role that HEB plays in allopolyploid invertebrate genomes. If HEB is constrained by features of the parental genomes, then we predict repeated evolution of similar HEB patterns among hybrid genomes formed from the same parental lineages. To address this, we reconstructed the history of hybridization between the New Zealand stick insect genera Acanthoxyla and Clitarchus using a high-quality genome assembly from Clitarchus hookeri to call variants and phase alleles. These analyses revealed the formation of three independent diploid and triploid hybrid lineages between these genera. RNA sequencing revealed a similar magnitude and direction of HEB among these hybrid lineages, and we observed that many enriched functions and pathways were also shared among lineages, consistent with repeated evolution due to parental genome constraints. In most hybrid lineages, a slight majority of the genes involved in mitochondrial function showed HEB towards the maternal homoeologues, consistent with only weak effects of mitonuclear incompatibility. We also observed a proteasome functional enrichment in most lineages and hypothesize this may result from the need to maintain proteostasis in hybrid genomes. Reference bias was a pervasive problem, and we caution against relying on HEB estimates from a single parental reference genome.
Keywords: Phasmatodea; asexual; gene expression; hybrid; transcriptomics.
© 2024 The Author(s). Molecular Ecology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2020 Jan 9;1(1):CD011535. doi: 10.1002/14651858.CD011535.pub3. Cochrane Database Syst Rev. 2020. Update in: Cochrane Database Syst Rev. 2021 Apr 19;4:CD011535. doi: 10.1002/14651858.CD011535.pub4. PMID: 31917873 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
-
Drugs for preventing postoperative nausea and vomiting in adults after general anaesthesia: a network meta-analysis.Cochrane Database Syst Rev. 2020 Oct 19;10(10):CD012859. doi: 10.1002/14651858.CD012859.pub2. Cochrane Database Syst Rev. 2020. PMID: 33075160 Free PMC article.
-
Sexual Harassment and Prevention Training.2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 36508513 Free Books & Documents.
References
-
- Adams, K. L. , Cronn, R. , Percifield, R. , & Wendel, J. F. (2003). Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ‐specific reciprocal silencing. Proceedings of the National Academy of Sciences of the United States of America, 100(8), 4649–4654. 10.1073/pnas.0630618100 - DOI - PMC - PubMed
-
- Andrews, S. (2010). FastQC: A quality control tool for high throughput sequence data . http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

