ATG10S promotes IFNL1 expression and autophagic degradation of multiple viral proteins mediated by IFNL1
- PMID: 38842055
- PMCID: PMC11423677
- DOI: 10.1080/15548627.2024.2361580
ATG10S promotes IFNL1 expression and autophagic degradation of multiple viral proteins mediated by IFNL1
Abstract
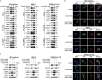
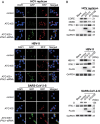
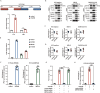
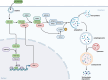
ATG10S is a newly discovered subtype of the autophagy protein ATG10. It promotes complete macroautophagy/autophagy, degrades multiple viral proteins, and increases the expression of type III interferons. Here, we aimed to investigate the mechanism of ATG10S cooperation with IFNL1 to degrade viral proteins from different viruses. Using western blot, immunoprecipitation (IP), tandem sensor RFP-GFP-LC3B and in situ proximity ligation assays, we showed that exogenous recombinant ATG10S protein (rHsATG10S) could enter into cells through clathrin, and ATG10S combined with ATG7 with IFNL1 assistance to facilitate ATG12-ATG5 conjugation, thereby contributing to the autophagosome formation in multiple cell lines containing different virions or viral proteins. The results of DNA IP and luciferase assays also showed that ATG10S was able to directly bind to a core motif (CAAGGG) within a binding site of transcription factor ZNF460 on the IFNL1 promoter, by which IFNL1 transcription was activated. These results clarified that ATG10S promoted autophagosome formation with the assistance of IFNL1 to ensure autophagy flux and autophagic degradation of multiple viral proteins and that ATG10S could also act as a novel transcription factor to promote IFNL1 gene expression. Importantly, this study further explored the antiviral mechanism of ATG10S interaction with type III interferon and provided a theoretical basis for the development of ATG10S into a new broad-spectrum antiviral protein drug.Abbreviation: ATG: autophagy related; ATG10S: the shorter isoform of autophagy-related 10; CC50: half cytotoxicity concentration; CCV: clathrin-coated transport vesicle; CLTC: clathrin heavy chain; CM: core motif; co-IP: co-immunoprecipitation; CPZ: chlorpromazine; ER: endoplasmic reticulum; HCV: hepatitis C virus; HBV: hepatitis B virus; HsCoV-OC43: Human coronavirus OC43; IFN: interferon; PLA: proximity ligation assay; rHsATG10S: recombinant human ATG10S protein; RLU: relative light unit; SQSTM1: sequestosome 1; ZNF: zinc finger protein.
Keywords: ATG10S; IFNL1 expression; ZNF460; autophagic degradation; core motif; viral proteins.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
