Pharmacogenetic Testing or Therapeutic Drug Monitoring: A Quantitative Framework
- PMID: 38842789
- PMCID: PMC11222190
- DOI: 10.1007/s40262-024-01382-3
Pharmacogenetic Testing or Therapeutic Drug Monitoring: A Quantitative Framework
Abstract
Background: Pharmacogenetic profiling and therapeutic drug monitoring (TDM) have both been proposed to manage inter-individual variability (IIV) in drug exposure. However, determining the most effective approach for estimating exposure for a particular drug remains a challenge. This study aimed to quantitatively assess the circumstances in which pharmacogenetic profiling may outperform TDM in estimating drug exposure, under three sources of variability (IIV, inter-occasion variability [IOV], and residual unexplained variability [RUV]).
Methods: Pharmacokinetic models were selected from the literature corresponding to drugs for which pharmacogenetic profiling and TDM are both clinically considered approaches for dose individualization. The models were used to simulate relevant drug exposures (trough concentration or area under the curve [AUC]) under varying degrees of IIV, IOV, and RUV.
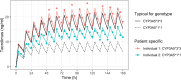
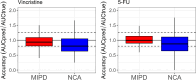
Results: Six drug cases were selected from the literature. Model-based simulations demonstrated that the percentage of patients for whom pharmacogenetic exposure prediction is superior to TDM differs for each drug case: tacrolimus (11.0%), tamoxifen (12.7%), efavirenz (49.2%), vincristine (49.6%), risperidone (48.1%), and 5-fluorouracil (5-FU) (100%). Generally, in the presence of higher unexplained IIV in combination with lower RUV and IOV, exposure was best estimated by TDM, whereas, under lower unexplained IIV in combination with higher IOV or RUV, pharmacogenetic profiling was preferred.
Conclusions: For the drugs with relatively low RUV and IOV (e.g., tamoxifen and tacrolimus), TDM estimated true exposure the best. Conversely, for drugs with similar or lower unexplained IIV (e.g., efavirenz or 5-FU, respectively) combined with relatively high RUV, pharmacogenetic profiling provided the most accurate estimate for most patients. However, genotype prevalence and the relative influence of genotypes on the PK, as well as the ability of TDM to accurately estimate AUC with a limited number of samples, had an impact. The results could be used to support clinical decision making when considering other factors, such as the probability for severe side effects.
© 2024. The Author(s).
Conflict of interest statement
MC, NR, AT, LEF, MOK, and LEF declare no competing interests for this work.
Figures

References
-
- Swen JJ, van der Wouden CH, Manson LE, Abdullah-Koolmees H, Blagec K, Blagus T, et al. A 12-gene pharmacogenetic panel to prevent adverse drug reactions: an open-label, multicentre, controlled, cluster-randomised crossover implementation study. Lancet (London, England). 2023;401:347–356. doi: 10.1016/S0140-6736(22)01841-4. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

