PIK3CA inhibition in models of proliferative glomerulonephritis and lupus nephritis
- PMID: 38842935
- PMCID: PMC11290976
- DOI: 10.1172/JCI176402
PIK3CA inhibition in models of proliferative glomerulonephritis and lupus nephritis
Abstract
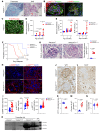
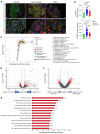
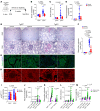
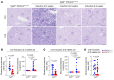
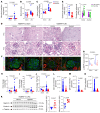
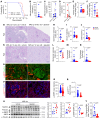
Proliferative glomerulonephritis is a severe condition that often leads to kidney failure. There is a significant lack of effective treatment for these disorders. Here, following the identification of a somatic PIK3CA gain-of-function mutation in podocytes of a patient, we demonstrate using multiple genetically engineered mouse models, single-cell RNA sequencing, and spatial transcriptomics the crucial role played by this pathway for proliferative glomerulonephritis development by promoting podocyte proliferation, dedifferentiation, and inflammation. Additionally, we show that alpelisib, a PI3Kα inhibitor, improves glomerular lesions and kidney function in different mouse models of proliferative glomerulonephritis and lupus nephritis by targeting podocytes. Surprisingly, we determined that pharmacological inhibition of PI3Kα affects B and T lymphocyte populations in lupus nephritis mouse models, with a decrease in the production of proinflammatory cytokines, autoantibodies, and glomerular complement deposition, which are all characteristic features of PI3Kδ inhibition, the primary PI3K isoform expressed in lymphocytes. Importantly, PI3Kα inhibition does not impact lymphocyte function under normal conditions. These findings were then confirmed in human lymphocytes isolated from patients with active lupus nephritis. In conclusion, we demonstrate the major role played by PI3Kα in proliferative glomerulonephritis and show that in this condition, alpelisib acts on both podocytes and the immune system.
Keywords: Molecular biology; Nephrology.
Conflict of interest statement
Figures





Comment in
- Genetic insights into the mechanisms of proliferative glomerulonephritis doi: 10.1172/JCI183090
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

