Harvesting and amplifying gene cassettes confers cross-resistance to critically important antibiotics
- PMID: 38843111
- PMCID: PMC11156391
- DOI: 10.1371/journal.ppat.1012235
Harvesting and amplifying gene cassettes confers cross-resistance to critically important antibiotics
Abstract
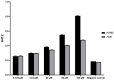
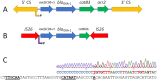
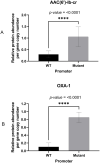
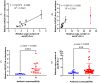
Amikacin and piperacillin/tazobactam are frequent antibiotic choices to treat bloodstream infection, which is commonly fatal and most often caused by bacteria from the family Enterobacterales. Here we show that two gene cassettes located side-by-side in and ancestral integron similar to In37 have been "harvested" by insertion sequence IS26 as a transposon that is widely disseminated among the Enterobacterales. This transposon encodes the enzymes AAC(6')-Ib-cr and OXA-1, reported, respectively, as amikacin and piperacillin/tazobactam resistance mechanisms. However, by studying bloodstream infection isolates from 769 patients from three hospitals serving a population of 1.2 million people in South West England, we show that increased enzyme production due to mutation in an IS26/In37-derived hybrid promoter or, more commonly, increased transposon copy number is required to simultaneously remove these two key therapeutic options; in many cases leaving only the last-resort antibiotic, meropenem. These findings may help improve the accuracy of predicting piperacillin/tazobactam treatment failure, allowing stratification of patients to receive meropenem or piperacillin/tazobactam, which may improve outcome and slow the emergence of meropenem resistance.
Copyright: © 2024 Dulyayangkul et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
In vitro activity of imipenem/relebactam against piperacillin/tazobactam-resistant and meropenem-resistant non-Morganellaceae Enterobacterales and Pseudomonas aeruginosa collected from patients with bloodstream, intra-abdominal and urinary tract infections in Western Europe: SMART 2018-2020.J Med Microbiol. 2023 Feb;72(2). doi: 10.1099/jmm.0.001645. J Med Microbiol. 2023. PMID: 36763081
-
Antimicrobial Activity of Ceftolozane-Tazobactam Tested Against Enterobacteriaceae and Pseudomonas aeruginosa with Various Resistance Patterns Isolated in U.S. Hospitals (2013-2016) as Part of the Surveillance Program: Program to Assess Ceftolozane-Tazobactam Susceptibility.Microb Drug Resist. 2018 Jun;24(5):563-577. doi: 10.1089/mdr.2017.0266. Epub 2017 Oct 17. Microb Drug Resist. 2018. PMID: 29039729
-
Antimicrobial Susceptibilities of Aerobic and Facultative Gram-Negative Bacilli from Intra-abdominal Infections in Patients from Seven Regions in China in 2012 and 2013.Antimicrob Agents Chemother. 2015 Oct 19;60(1):245-51. doi: 10.1128/AAC.00956-15. Print 2016 Jan. Antimicrob Agents Chemother. 2015. PMID: 26482308 Free PMC article.
-
Integrons in Enterobacteriaceae: diversity, distribution and epidemiology.Int J Antimicrob Agents. 2018 Feb;51(2):167-176. doi: 10.1016/j.ijantimicag.2017.10.004. Epub 2017 Oct 14. Int J Antimicrob Agents. 2018. PMID: 29038087 Review.
-
Optimizing therapy of bloodstream infection due to extended-spectrum β-lactamase-producing Enterobacteriaceae.Curr Opin Crit Care. 2019 Oct;25(5):438-448. doi: 10.1097/MCC.0000000000000646. Curr Opin Crit Care. 2019. PMID: 31369411 Review.
Cited by
-
Three simple and cost-effective assays for AAC(6')-Ib-cr enzyme activity.Front Microbiol. 2025 Apr 25;16:1513425. doi: 10.3389/fmicb.2025.1513425. eCollection 2025. Front Microbiol. 2025. PMID: 40351320 Free PMC article.
References
-
- Rudd KE, Johnson SC, Agesa KM, Shackelford KA, Tsoi D, Kievlan DR, Colombara DV, Ikuta KS, Kissoon N, Finfer S, Fleischmann-Struzek C, Machado FR, Reinhart KK, Rowan K, Seymour CW, Watson RS, West TE, Marinho F, Hay SI, Lozano R, Lopez AD, Angus DC, Murray CJL, Naghavi M. 2020. Global, regional, and national sepsis incidence and mortality, 1990–2017: analysis for the Global Burden of Disease Study. Lancet (London, England). 95:200–11. doi: 10.1016/S0140-6736(19)32989-7 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

