Genome-wide identification and characterization of DTX family genes highlighting their locations, functions, and regulatory factors in banana (Musa acuminata)
- PMID: 38843276
- PMCID: PMC11156367
- DOI: 10.1371/journal.pone.0303065
Genome-wide identification and characterization of DTX family genes highlighting their locations, functions, and regulatory factors in banana (Musa acuminata)
Abstract
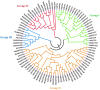
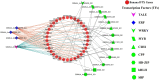
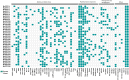
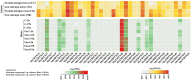
The detoxification efflux carriers (DTX) are a significant group of multidrug efflux transporter family members that play diverse functions in all kingdoms of living organisms. However, genome-wide identification and characterization of DTX family transporters have not yet been performed in banana, despite its importance as an economic fruit plant. Therefore, a detailed genome-wide analysis of DTX family transporters in banana (Musa acuminata) was conducted using integrated bioinformatics and systems biology approaches. In this study, a total of 37 DTX transporters were identified in the banana genome and divided into four groups (I, II, III, and IV) based on phylogenetic analysis. The gene structures, as well as their proteins' domains and motifs, were found to be significantly conserved. Gene ontology (GO) annotation revealed that the predicted DTX genes might play a vital role in protecting cells and membrane-bound organelles through detoxification mechanisms and the removal of drug molecules from banana cells. Gene regulatory analyses identified key transcription factors (TFs), cis-acting elements, and post-transcriptional regulators (miRNAs) of DTX genes, suggesting their potential roles in banana. Furthermore, the changes in gene expression levels due to pathogenic infections and non-living factor indicate that banana DTX genes play a role in responses to both biotic and abiotic stresses. The results of this study could serve as valuable tools to improve banana quality by protecting them from a range of environmental stresses.
Copyright: © 2024 Amin et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







Similar articles
-
Genome-wide identification of DCL, AGO and RDR gene families and their associated functional regulatory elements analyses in banana (Musa acuminata).PLoS One. 2021 Sep 2;16(9):e0256873. doi: 10.1371/journal.pone.0256873. eCollection 2021. PLoS One. 2021. PMID: 34473743 Free PMC article.
-
Genome-wide identification and characterization of the superoxide dismutase gene family in Musa acuminata cv. Tianbaojiao (AAA group).BMC Genomics. 2015 Oct 20;16:823. doi: 10.1186/s12864-015-2046-7. BMC Genomics. 2015. PMID: 26486759 Free PMC article.
-
Genome-Wide Identification and Expression Analysis of the TGA Gene Family in Banana (Musa nana Lour.) Under Various Nitrogen Conditions.Int J Mol Sci. 2025 Feb 28;26(5):2168. doi: 10.3390/ijms26052168. Int J Mol Sci. 2025. PMID: 40076789 Free PMC article.
-
Genome-wide identification, evolution analysis of LysM gene family members and their expression analysis in response to biotic and abiotic stresses in banana (Musa L.).Gene. 2022 Dec 15;845:146849. doi: 10.1016/j.gene.2022.146849. Epub 2022 Aug 28. Gene. 2022. PMID: 36044944
-
Genome-Wide Identification and Analysis of U-Box E3 Ubiquitin⁻Protein Ligase Gene Family in Banana.Int J Mol Sci. 2018 Dec 4;19(12):3874. doi: 10.3390/ijms19123874. Int J Mol Sci. 2018. PMID: 30518127 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

