Temporal regulation of MDA5 inactivation by Caspase-3 dependent cleavage of 14-3-3η
- PMID: 38843304
- PMCID: PMC11185488
- DOI: 10.1371/journal.ppat.1012287
Temporal regulation of MDA5 inactivation by Caspase-3 dependent cleavage of 14-3-3η
Abstract
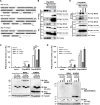
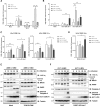
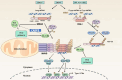
The kinetics of type I interferon (IFN) induction versus the virus replication compete, and the result of the competition determines the outcome of the infection. Chaperone proteins that involved in promoting the activation kinetics of PRRs rapidly trigger antiviral innate immunity. We have previously shown that prior to the interaction with MAVS to induce type I IFN, 14-3-3η facilitates the oligomerization and intracellular redistribution of activated MDA5. Here we report that the cleavage of 14-3-3η upon MDA5 activation, and we identified Caspase-3 activated by MDA5-dependent signaling was essential to produce sub-14-3-3η lacking the C-terminal helix (αI) and tail. The cleaved form of 14-3-3η (sub-14-3-3η) could strongly interact with MDA5 but could not support MDA5-dependent type I IFN induction, indicating the opposite functions between the full-length 14-3-3η and sub-14-3-3η. During human coronavirus or enterovirus infections, the accumulation of sub-14-3-3η was observed along with the activation of Caspase-3, suggesting that RNA viruses may antagonize 14-3-3η by promoting the formation of sub-14-3-3η to impair antiviral innate immunity. In conclusion, sub-14-3-3η, which could not promote MDA5 activation, may serve as a negative feedback to return to homeostasis to prevent excessive type I IFN production and unnecessary inflammation.
Copyright: © 2024 Chan et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
The 14-3-3η chaperone protein promotes antiviral innate immunity via facilitating MDA5 oligomerization and intracellular redistribution.PLoS Pathog. 2019 Feb 11;15(2):e1007582. doi: 10.1371/journal.ppat.1007582. eCollection 2019 Feb. PLoS Pathog. 2019. PMID: 30742689 Free PMC article.
-
Role of Enteroviral RNA-Dependent RNA Polymerase in Regulation of MDA5-Mediated Beta Interferon Activation.J Virol. 2019 May 1;93(10):e00132-19. doi: 10.1128/JVI.00132-19. Print 2019 May 15. J Virol. 2019. PMID: 30814289 Free PMC article.
-
DNAJB1/HSP40 Suppresses Melanoma Differentiation-Associated Gene 5-Mitochondrial Antiviral Signaling Protein Function in Conjunction with HSP70.J Innate Immun. 2018;10(1):44-55. doi: 10.1159/000480740. Epub 2017 Oct 26. J Innate Immun. 2018. PMID: 29069650 Free PMC article.
-
MDA5 Is a Major Determinant of Developing Symptoms in Critically Ill COVID-19 Patients.Clin Rev Allergy Immunol. 2024 Dec;67(1-3):58-72. doi: 10.1007/s12016-024-09008-z. Epub 2024 Oct 26. Clin Rev Allergy Immunol. 2024. PMID: 39460899 Review.
-
A Balancing Act: MDA5 in Antiviral Immunity and Autoinflammation.Trends Microbiol. 2019 Jan;27(1):75-85. doi: 10.1016/j.tim.2018.08.007. Epub 2018 Sep 7. Trends Microbiol. 2019. PMID: 30201512 Free PMC article. Review.
Cited by
-
How the Structure of Signaling Regulation Evolves: Insights From an Evolutionary Model.Mol Biol Evol. 2025 Apr 30;42(5):msaf104. doi: 10.1093/molbev/msaf104. Mol Biol Evol. 2025. PMID: 40341936 Free PMC article.
-
RNF5 inhibits HBV replication by mediating caspase-3-dependent degradation of core protein.Front Microbiol. 2025 Apr 1;16:1548061. doi: 10.3389/fmicb.2025.1548061. eCollection 2025. Front Microbiol. 2025. PMID: 40236486 Free PMC article.
-
How the Structure of Signaling Regulation Evolves: Insights from an Evolutionary Model.bioRxiv [Preprint]. 2024 Oct 25:2024.10.23.619883. doi: 10.1101/2024.10.23.619883. bioRxiv. 2024. Update in: Mol Biol Evol. 2025 Apr 30;42(5):msaf104. doi: 10.1093/molbev/msaf104. PMID: 39484560 Free PMC article. Updated. Preprint.
References
-
- Breiman A, Grandvaux N, Lin R, Ottone C, Akira S, Yoneyama M, et al.. Inhibition of RIG-I-dependent signaling to the interferon pathway during hepatitis C virus expression and restoration of signaling by IKKepsilon. Journal of virology. 2005;79(7):3969–78. Epub 2005/03/16. doi: 10.1128/JVI.79.7.3969-3978.2005 [pii] ; PubMed Central PMCID: PMC1061556. - DOI - PMC - PubMed
-
- Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, et al.. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(2):582–7. doi: 10.1073/pnas.0606699104 ; PubMed Central PMCID: PMC1766428. - DOI - PMC - PubMed
-
- Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM, et al.. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(15):6001–6. Epub 2006/04/06. doi: 10.1073/pnas.0601523103 [pii] ; PubMed Central PMCID: PMC1458687. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

