Cell Culture Differentiation and Proliferation Conditions Influence the In Vitro Regeneration of the Human Airway Epithelium
- PMID: 38843491
- PMCID: PMC11376247
- DOI: 10.1165/rcmb.2023-0356MA
Cell Culture Differentiation and Proliferation Conditions Influence the In Vitro Regeneration of the Human Airway Epithelium
Abstract
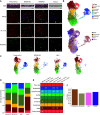
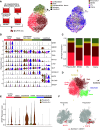
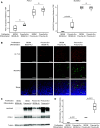
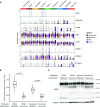
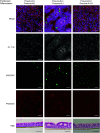
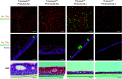
The human airway mucociliary epithelium can be recapitulated in vitro using primary cells cultured in an air-liquid interface (ALI), a reliable surrogate to perform pathophysiological studies. As tremendous variations exist among media used for ALI-cultured human airway epithelial cells, the aim of our study was to evaluate the impact of several media (BEGM, PneumaCult, Half & Half, and Clancy) on cell type distribution using single-cell RNA sequencing and imaging. Our work revealed the impact of these media on cell composition, gene expression profile, cell signaling, and epithelial morphology. We found higher proportions of multiciliated cells in PneumaCult-ALI and Half & Half, stronger EGF signaling from basal cells in BEGM-ALI, differential expression of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) entry factor ACE2, and distinct secretome transcripts depending on the media used. We also established that proliferation in PneumaCult-Ex Plus favored secretory cell fate, showing the key influence of proliferation media on late differentiation epithelial characteristics. Altogether, our data offer a comprehensive repertoire for evaluating the effects of culture conditions on airway epithelial differentiation and will aid in choosing the most relevant medium according to the processes to be investigated, such as cilia, mucus biology, or viral infection. We detail useful parameters that should be explored to document airway epithelial cell fate and morphology.
Keywords: airway epithelium; air–liquid interface cell culture; culture medium; differentiation; single-cell RNA sequencing.
Figures

References
-
- Deprez M, Zaragosi L-E, Truchi M, Bécavin C, Ruiz García S, Arguel MJ, et al. A single-cell atlas of the human healthy airways. Am J Respir Crit Care Med . 2020;202:1636–1645. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

