Biliverdin Reductase-A integrates insulin signaling with mitochondrial metabolism through phosphorylation of GSK3β
- PMID: 38843768
- PMCID: PMC11190564
- DOI: 10.1016/j.redox.2024.103221
Biliverdin Reductase-A integrates insulin signaling with mitochondrial metabolism through phosphorylation of GSK3β
Abstract
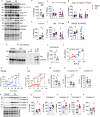
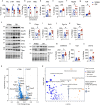
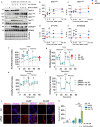
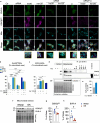
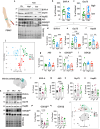
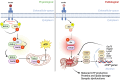
Brain insulin resistance links the failure of energy metabolism with cognitive decline in both type 2 Diabetes Mellitus (T2D) and Alzheimer's disease (AD), although the molecular changes preceding overt brain insulin resistance remain unexplored. Abnormal biliverdin reductase-A (BVR-A) levels were observed in both T2D and AD and were associated with insulin resistance. Here, we demonstrate that reduced BVR-A levels alter insulin signaling and mitochondrial bioenergetics in the brain. Loss of BVR-A leads to IRS1 hyper-activation but dysregulates Akt-GSK3β complex in response to insulin, hindering the accumulation of pGSK3βS9 into the mitochondria. This event impairs oxidative phosphorylation and fosters the activation of the mitochondrial Unfolded Protein Response (UPRmt). Remarkably, we unveil that BVR-A is required to shuttle pGSK3βS9 into the mitochondria. Our data sheds light on the intricate interplay between insulin signaling and mitochondrial metabolism in the brain unraveling potential targets for mitigating the development of brain insulin resistance and neurodegeneration.
Keywords: Biliverdin reductase-A; Brain insulin resistance; GSK3β; Mitochondrial metabolism; Mitochondrial unfolded protein response; Oxidative stress.
Copyright © 2024 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- Biessels G.J., Koffeman A., Scheltens P. Diabetes and cognitive impairment. Clinical diagnosis and brain imaging in patients attending a memory clinic. J. Neurol. 2006;253(4):477–482. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

