Widespread horse-based mobility arose around 2200 BCE in Eurasia
- PMID: 38843826
- PMCID: PMC11269178
- DOI: 10.1038/s41586-024-07597-5
Widespread horse-based mobility arose around 2200 BCE in Eurasia
Abstract
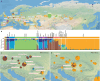
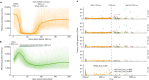
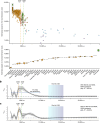
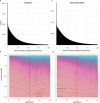
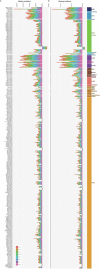
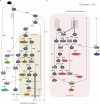
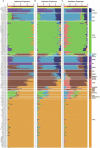
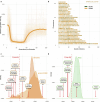
Horses revolutionized human history with fast mobility1. However, the timeline between their domestication and their widespread integration as a means of transport remains contentious2-4. Here we assemble a collection of 475 ancient horse genomes to assess the period when these animals were first reshaped by human agency in Eurasia. We find that reproductive control of the modern domestic lineage emerged around 2200 BCE, through close-kin mating and shortened generation times. Reproductive control emerged following a severe domestication bottleneck starting no earlier than approximately 2700 BCE, and coincided with a sudden expansion across Eurasia that ultimately resulted in the replacement of nearly every local horse lineage. This expansion marked the rise of widespread horse-based mobility in human history, which refutes the commonly held narrative of large horse herds accompanying the massive migration of steppe peoples across Europe around 3000 BCE and earlier3,5. Finally, we detect significantly shortened generation times at Botai around 3500 BCE, a settlement from central Asia associated with corrals and a subsistence economy centred on horses6,7. This supports local horse husbandry before the rise of modern domestic bloodlines.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Kelekna, P. The Horse in Human History (Cambridge Univ. Press, 2009).
-
- Anthony, D. W. The Horse, the Wheel, and Language: How Bronze-Age Riders from the Eurasian Steppes Shaped the Modern World (Princeton Univ. Press, 2007).

