Adipose-derived mesenchymal stem cells' adipogenesis chemistry analyzed by FTIR and Raman metrics
- PMID: 38844049
- PMCID: PMC11260339
- DOI: 10.1016/j.jlr.2024.100573
Adipose-derived mesenchymal stem cells' adipogenesis chemistry analyzed by FTIR and Raman metrics
Abstract
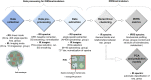
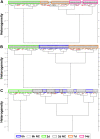
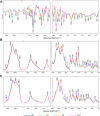
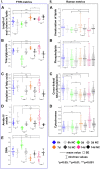
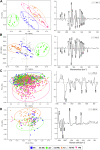
The full understanding of molecular mechanisms of cell differentiation requires a holistic view. Here we combine label-free FTIR and Raman hyperspectral imaging with data mining to detect the molecular cell composition enabling noninvasive monitoring of cell differentiation and identifying biochemical heterogeneity. Mouse adipose-derived mesenchymal stem cells (AD-MSCs) undergoing adipogenesis were followed by Raman and FT-IR imaging, Oil Red, and immunofluorescence. A workflow of the data analysis (IRRSmetrics4stem) was designed to identify spectral predictors of adipogenesis and test machine-learning (ML) methods (hierarchical clustering, PCA, PLSR) for the control of the AD-MSCs differentiation degree. IRRSmetrics4stem provided insights into the chemism of adipogenesis. With single-cell tracking, we established IRRS metrics for lipids, proteins, and DNA variations during AD-MSCs differentiation. The over 90% predictive efficiency of the selected ML methods proved the high sensitivity of the IRRS metrics. Importantly, the IRRS metrics unequivocally recognize a switch from proliferation to differentiation. This study introduced a new bioassay identifying molecular markers indicating molecular transformations and delivering rapid and machine learning-based monitoring of adipogenesis that can be relevant to other differentiation processes. Thus, we introduce a novel, rapid, machine learning-based bioassay to identify molecular markers of adipogenesis. It can be relevant to identification of differentiation-related molecular processes in other cell types, and beyond the cell differentiation including progression of different cellular pathophysiologies reconstituted in vitro.
Keywords: adipogenesis; mesenchymal stem cells; prediction of differentiation stage; regenerative medicine; spectroscopic molecular imaging.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Alenzi F.Q.B., Bahkali A.H. Stem cells: biology and clinical potential. Afr. J. Biotechnol. 2011;10:19929–19940.
-
- Keung E., Nelson P., Conrad C. Concise review: adipose-derived stem cells as a novel tool for future regenerative medicine. Stem Cells. 2013;30:804–810. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

