Exploration of mobile genetic elements in the ruminal microbiome of Nellore cattle
- PMID: 38844487
- PMCID: PMC11156634
- DOI: 10.1038/s41598-024-63951-7
Exploration of mobile genetic elements in the ruminal microbiome of Nellore cattle
Abstract
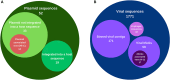
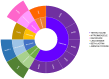
Metagenomics has made it feasible to elucidate the intricacies of the ruminal microbiome and its role in the differentiation of animal production phenotypes of significance. The search for mobile genetic elements (MGEs) has taken on great importance, as they play a critical role in the transfer of genetic material between organisms. Furthermore, these elements serve a dual purpose by controlling populations through lytic bacteriophages, thereby maintaining ecological equilibrium and driving the evolutionary progress of host microorganisms. In this study, we aimed to identify the association between ruminal bacteria and their MGEs in Nellore cattle using physical chromosomal links through the Hi-C method. Shotgun metagenomic sequencing and the proximity ligation method ProxiMeta were used to analyze DNA, getting 1,713,111,307 bp, which gave rise to 107 metagenome-assembled genomes from rumen samples of four Nellore cows maintained on pasture. Taxonomic analysis revealed that most of the bacterial genomes belonged to the families Lachnospiraceae, Bacteroidaceae, Ruminococcaceae, Saccharofermentanaceae, and Treponemataceae and mostly encoded pathways for central carbon and other carbohydrate metabolisms. A total of 31 associations between host bacteria and MGE were identified, including 17 links to viruses and 14 links to plasmids. Additionally, we found 12 antibiotic resistance genes. To our knowledge, this is the first study in Brazilian cattle that connect MGEs with their microbial hosts. It identifies MGEs present in the rumen of pasture-raised Nellore cattle, offering insights that could advance biotechnology for food digestion and improve ruminant performance in production systems.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- González-Recio O, et al. Invited review: Novel methods and perspectives for modulating the rumen microbiome through selective breeding as a means to improve complex traits: Implications for methane emissions in cattle. Livest. Sci. 2023;269:105171. doi: 10.1016/j.livsci.2023.105171. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

