Nucleotide metabolism in cancer cells fuels a UDP-driven macrophage cross-talk, promoting immunosuppression and immunotherapy resistance
- PMID: 38844817
- PMCID: PMC11358017
- DOI: 10.1038/s43018-024-00771-8
Nucleotide metabolism in cancer cells fuels a UDP-driven macrophage cross-talk, promoting immunosuppression and immunotherapy resistance
Abstract
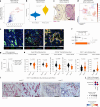
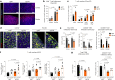
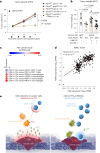
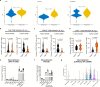
Many individuals with cancer are resistant to immunotherapies. Here, we identify the gene encoding the pyrimidine salvage pathway enzyme cytidine deaminase (CDA) among the top upregulated metabolic genes in several immunotherapy-resistant tumors. We show that CDA in cancer cells contributes to the uridine diphosphate (UDP) pool. Extracellular UDP hijacks immunosuppressive tumor-associated macrophages (TAMs) through its receptor P2Y6. Pharmacologic or genetic inhibition of CDA in cancer cells (or P2Y6 in TAMs) disrupts TAM-mediated immunosuppression, promoting cytotoxic T cell entry and susceptibility to anti-programmed cell death protein 1 (anti-PD-1) treatment in resistant pancreatic ductal adenocarcinoma (PDAC) and melanoma models. Conversely, CDA overexpression in CDA-depleted PDACs or anti-PD-1-responsive colorectal tumors or systemic UDP administration (re)establishes resistance. In individuals with PDAC, high CDA levels in cancer cells correlate with increased TAMs, lower cytotoxic T cells and possibly anti-PD-1 resistance. In a pan-cancer single-cell atlas, CDAhigh cancer cells match with T cell cytotoxicity dysfunction and P2RY6high TAMs. Overall, we suggest CDA and P2Y6 as potential targets for cancer immunotherapy.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no conflicts of interest.
Figures










References
-
- Smyth, M. J., Ngiow, S. F., Ribas, A. & Teng, M. W. Combination cancer immunotherapies tailored to the tumour microenvironment. Nat. Rev. Clin. Oncol.13, 143–158 (2015). - PubMed
-
- Herbst, R. S. et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet387, 1540–1550 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- G0B4620N/Fonds Wetenschappelijk Onderzoek (Research Foundation Flanders)
- 28245/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
- G0A7419N/Fonds Wetenschappelijk Onderzoek (Research Foundation Flanders)
- OT2 OD030544/OD/NIH HHS/United States
- 11M9922N/Fonds Wetenschappelijk Onderzoek (Research Foundation Flanders)
- SEG9251/European Molecular Biology Organization (EMBO)
- ImmunoFit, 773208/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- U2C DK119886/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

