Genome-wide identification and expression analysis of the KNOX family and its diverse roles in response to growth and abiotic tolerance in sweet potato and its two diploid relatives
- PMID: 38844832
- PMCID: PMC11157901
- DOI: 10.1186/s12864-024-10470-4
Genome-wide identification and expression analysis of the KNOX family and its diverse roles in response to growth and abiotic tolerance in sweet potato and its two diploid relatives
Abstract
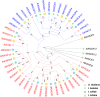
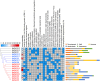
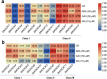
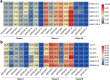
KNOXs, a type of homeobox genes that encode atypical homeobox proteins, play an essential role in the regulation of growth and development, hormonal response, and abiotic stress in plants. However, the KNOX gene family has not been explored in sweet potato. In this study, through sequence alignment, genomic structure analysis, and phylogenetic characterization, 17, 12 and 11 KNOXs in sweet potato (I. batatas, 2n = 6x = 90) and its two diploid relatives I. trifida (2n = 2x = 30) and I. triloba (2n = 2x = 30) were identified. The protein physicochemical properties, chromosome localization, phylogenetic relationships, gene structure, protein interaction network, cis-elements of promoters, tissue-specific expression and expression patterns under hormone treatment and abiotic stresses of these 40 KNOX genes were systematically studied. IbKNOX4, -5, and - 6 were highly expressed in the leaves of the high-yield varieties Longshu9 and Xushu18. IbKNOX3 and IbKNOX8 in Class I were upregulated in initial storage roots compared to fibrous roots. IbKNOXs in Class M were specifically expressed in the stem tip and hardly expressed in other tissues. Moreover, IbKNOX2 and - 6, and their homologous genes were induced by PEG/mannitol and NaCl treatments. The results showed that KNOXs were involved in regulating growth and development, hormone crosstalk and abiotic stress responses between sweet potato and its two diploid relatives. This study provides a comparison of these KNOX genes in sweet potato and its two diploid relatives and a theoretical basis for functional studies.
Keywords: I. trifida; I. triloba; KNOX; Abiotic stress; Hormone treatment; Sweet potato; Tissue-specific expression.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- Song WH, Dong TT, Yu JW, Tan CT, Yu YH, Li ZY. The transcription factors involved in storage root/stem formation and thickening of tuber crops. Mol Plant Breed. 2017;15(5):1708–17.
-
- Ye SG, Zai WS, Xiong ZL, Zhang HL, Ma YR. Genome-wide identification of KNOX gene family in tomato and their evolutionary relationship in Solanaceae. Acta Agric Nucl Sin. 2017;31(7):1263–71. doi: 10.11869/j.issn.100-8551.2017.07.1263. - DOI
MeSH terms
Substances
Grants and funding
- CARS-10-Sweetpotato/the earmarked fund for CARS-10-Sweetpotato
- CARS-10-Sweetpotato/the earmarked fund for CARS-10-Sweetpotato
- CARS-10-Sweetpotato/the earmarked fund for CARS-10-Sweetpotato
- CARS-10-Sweetpotato/the earmarked fund for CARS-10-Sweetpotato
- CARS-10-Sweetpotato/the earmarked fund for CARS-10-Sweetpotato
- CARS-10-Sweetpotato/the earmarked fund for CARS-10-Sweetpotato
- CARS-10-Sweetpotato/the earmarked fund for CARS-10-Sweetpotato
- 2023YFD1200700/2023YFD1200703/the National Key Research and Development Program of China
- 2023YFD1200700/2023YFD1200703/the National Key Research and Development Program of China
- 2023YFD1200700/2023YFD1200703/the National Key Research and Development Program of China
- 2023YFD1200700/2023YFD1200703/the National Key Research and Development Program of China
- SCKJ-JYRC-2022-61/SYND-2022-09/the Project of Sanya Yazhou Bay Science and Technology City
- SCKJ-JYRC-2022-61/SYND-2022-09/the Project of Sanya Yazhou Bay Science and Technology City
- SCKJ-JYRC-2022-61/SYND-2022-09/the Project of Sanya Yazhou Bay Science and Technology City
- 6212017/the Beijing Natural Science Foundation
- 6212017/the Beijing Natural Science Foundation
- 6212017/the Beijing Natural Science Foundation
LinkOut - more resources
Full Text Sources
Research Materials

