Comparative transcriptomic analysis primarily explores the molecular mechanism of compound eye formation in Neocaridina denticulata sinensis
- PMID: 38844864
- PMCID: PMC11155044
- DOI: 10.1186/s12864-024-10453-5
Comparative transcriptomic analysis primarily explores the molecular mechanism of compound eye formation in Neocaridina denticulata sinensis
Abstract
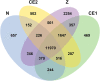
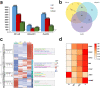
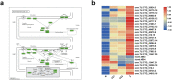
Compound eyes formation in decapod crustaceans occurs after the nauplius stage. However, the key genes and regulatory mechanisms of compound eye development during crustacean embryonic development have not yet been clarified. In this study, RNA-seq was used to investigate the gene expression profiles of Neocaridina denticulata sinensis from nauplius to zoea stage. Based on RNA-seq data analysis, the phototransduction and insect hormone biosynthesis pathways were enriched, and molting-related neuropeptides were highly expressed. There was strong cell proliferation in the embryo prior to compound eye development. The formation of the visual system and the hormonal regulation of hatching were the dominant biological events during compound eye development. The functional analysis of DEGs across all four developmental stages showed that cuticle formation, muscle growth and the establishment of immune system occurred from nauplius to zoea stage. Key genes related to eye development were discovered, including those involved in the determination and differentiation of the eye field, eye-color formation, and visual signal transduction. In conclusion, the results increase the understanding of the molecular mechanism of eye formation in crustacean embryonic stage.
Keywords: Neocaridina denticulata sinensis; Comparative transcriptomic analysis; Compound eye; Differentially expressed genes; Embryonic development.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
There is no conflict of interest.
Figures





Similar articles
-
Functional analysis of ABCG2 gene in pigment transport of Neocaridina denticulata sinensis.Gene. 2022 Nov 30;844:146810. doi: 10.1016/j.gene.2022.146810. Epub 2022 Aug 17. Gene. 2022. PMID: 35985411
-
Genome-wide identification and expression profiling of Wnt gene family in Neocaridina denticulata sinensis.Gene. 2023 Feb 20;854:147122. doi: 10.1016/j.gene.2022.147122. Epub 2022 Dec 17. Gene. 2023. PMID: 36539046
-
RNA-seq analysis revealing the immune response of Neocaridina denticulata sinensis gill to Vibrio parahaemolyticus infection.Fish Shellfish Immunol. 2022 Nov;130:409-417. doi: 10.1016/j.fsi.2022.09.049. Epub 2022 Sep 23. Fish Shellfish Immunol. 2022. PMID: 36154891
-
Genetic and developmental mechanisms underlying the formation of the Drosophila compound eye.Dev Dyn. 2012 Jan;241(1):40-56. doi: 10.1002/dvdy.22738. Epub 2011 Sep 19. Dev Dyn. 2012. PMID: 21932322 Review.
-
Crustacean sexual differentiation: a decapod perspective.Curr Opin Insect Sci. 2025 Jun;69:101371. doi: 10.1016/j.cois.2025.101371. Epub 2025 Mar 23. Curr Opin Insect Sci. 2025. PMID: 40132748 Review.
Cited by
-
Integrating multi-omics data to identify the role of Aggrephagy-related genes in tumor microenvironment and key tumorigenesis factors of GB from the perspective of single-cell sequencing.Discov Oncol. 2025 May 16;16(1):777. doi: 10.1007/s12672-025-02431-4. Discov Oncol. 2025. PMID: 40377747 Free PMC article.
References
-
- CW O, CW M, RG H, HL S. Reproduction and population dynamics of the temperate freshwater shrimp, Neocaridina denticulata denticulata (De Haan, 1844), in a Korean stream. Crustaceana. 2003;76:993–1015. doi: 10.1163/156854003771997864. - DOI
-
- Nur FAH, Christianu A. Breeding and life cycle of Neocaridina denticulata sinensis (Kemp, 1918) Asian J Anim Vet Adv. 2012;8(1):108–15. doi: 10.3923/ajava.2013.108.115. - DOI
-
- Sun S, Fan C, Li F, Mu S, Tang H, Zhang H, Kang X. Preliminary study on the conformation development of Neocaridina denticulate sinensis. Hebei Fisheries 2007(12):22–5 (in Chinese).
-
- Cao L, Qin Z, Jiang Y, Liu X, Li X, Lin Y, Huang K, Liu X. Embryonic development of Caridina Japonica and in vitro incubation of its fertilized eggs. Progress Fish Sci. 2020;41(1):145–52.
Publication types
MeSH terms
Grants and funding
- D2022201003/Natural Science Foundation of Hebei Province of China
- D2023201002/Natural Science Foundation of Hebei Province of China
- 32172954/National Natural Science Foundation of China
- ZD2022093/Science Research Project of Hebei Education Department
- 22323201D/Key Research and Development Project of Hebei Province
LinkOut - more resources
Full Text Sources

