NSUN2/YBX1 promotes the progression of breast cancer by enhancing HGH1 mRNA stability through m5C methylation
- PMID: 38844963
- PMCID: PMC11155144
- DOI: 10.1186/s13058-024-01847-0
NSUN2/YBX1 promotes the progression of breast cancer by enhancing HGH1 mRNA stability through m5C methylation
Abstract
Background: RNA m5C methylation has been extensively implicated in the occurrence and development of tumors. As the main methyltransferase, NSUN2 plays a crucial regulatory role across diverse tumor types. However, the precise impact of NSUN2-mediated m5C modification on breast cancer (BC) remains unclear. Our study aims to elucidate the molecular mechanism underlying how NSUN2 regulates the target gene HGH1 (also known as FAM203) through m5C modification, thereby promoting BC progression. Additionally, this study targets at preliminarily clarifying the biological roles of NSUN2 and HGH1 in BC.
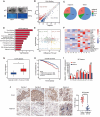
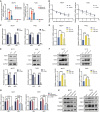
Methods: Tumor and adjacent tissues from 5 BC patients were collected, and the m5C modification target HGH1 in BC was screened through RNA sequencing (RNA-seq) and single-base resolution m5C methylation sequencing (RNA-BisSeq). Methylation RNA immunoprecipitation-qPCR (MeRIP-qPCR) and RNA-binding protein immunoprecipitation-qPCR (RIP-qPCR) confirmed that the methylation molecules NSUN2 and YBX1 specifically recognized and bound to HGH1 through m5C modification. In addition, proteomics, co-immunoprecipitation (co-IP), and Ribosome sequencing (Ribo-Seq) were used to explore the biological role of HGH1 in BC.
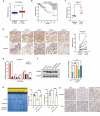
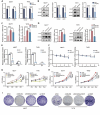
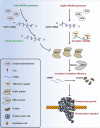
Results: As the main m5C methylation molecule, NSUN2 is abnormally overexpressed in BC and increases the overall level of RNA m5C. Knocking down NSUN2 can inhibit BC progression in vitro or in vivo. Combined RNA-seq and RNA-BisSeq analysis identified HGH1 as a potential target of abnormal m5C modifications. We clarified the mechanism by which NSUN2 regulates HGH1 expression through m5C modification, a process that involves interactions with the YBX1 protein, which collectively impacts mRNA stability and protein synthesis. Furthermore, this study is the first to reveal the binding interaction between HGH1 and the translation elongation factor EEF2, providing a comprehensive understanding of its ability to regulate transcript translation efficiency and protein synthesis in BC cells.
Conclusions: This study preliminarily clarifies the regulatory role of the NSUN2-YBX1-m5C-HGH1 axis from post-transcriptional modification to protein translation, revealing the key role of abnormal RNA m5C modification in BC and suggesting that HGH1 may be a new epigenetic biomarker and potential therapeutic target for BC.
Keywords: Breast cancer; HGH1; NSUN2; RNA 5-methylcytosinine; Translation efficiency.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- NICE, Early and locally advanced breast cancer: diagnosis and management, in Early and locally advanced breast cancer: diagnosis and management. 2024: London.
Publication types
MeSH terms
Substances
Grants and funding
- LHGI20230280/The Henan Medical Science and Technology Research Plan Provincial Project
- LHGI20230280/The Henan Medical Science and Technology Research Plan Provincial Project
- LHGI20230280/The Henan Medical Science and Technology Research Plan Provincial Project
- ZYCXTD2023010/The Funding for Scientific Research and Innovation Team of The First Affiliated Hospital of Zhengzhou University
- ZYCXTD2023010/The Funding for Scientific Research and Innovation Team of The First Affiliated Hospital of Zhengzhou University
- ZYCXTD2023010/The Funding for Scientific Research and Innovation Team of The First Affiliated Hospital of Zhengzhou University
- ZYCXTD2023010/The Funding for Scientific Research and Innovation Team of The First Affiliated Hospital of Zhengzhou University
- ZYCXTD2023010/The Funding for Scientific Research and Innovation Team of The First Affiliated Hospital of Zhengzhou University
- ZYCXTD2023010/The Funding for Scientific Research and Innovation Team of The First Affiliated Hospital of Zhengzhou University
- ZYCXTD2023010/The Funding for Scientific Research and Innovation Team of The First Affiliated Hospital of Zhengzhou University
- HNSWJW-2020017/The Special Fund for Young and Middle School Leaders of Henan Health Commission
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

