Genetic analysis of 106 sporadic cases with hearing loss in the UAE population
- PMID: 38844983
- PMCID: PMC11157727
- DOI: 10.1186/s40246-024-00630-8
Genetic analysis of 106 sporadic cases with hearing loss in the UAE population
Abstract
Background: Hereditary hearing loss is a rare hereditary condition that has a significant presence in consanguineous populations. Despite its prevalence, hearing loss is marked by substantial genetic diversity, which poses challenges for diagnosis and screening, particularly in cases with no clear family history or when the impact of the genetic variant requires functional analysis, such as in the case of missense mutations and UTR variants. The advent of next-generation sequencing (NGS) has transformed the identification of genes and variants linked to various conditions, including hearing loss. However, there remains a high proportion of undiagnosed patients, attributable to various factors, including limitations in sequencing coverage and gaps in our knowledge of the entire genome, among other factors. In this study, our objective was to comprehensively identify the spectrum of genes and variants associated with hearing loss in a cohort of 106 affected individuals from the UAE.
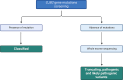
Results: In this study, we investigated 106 sporadic cases of hearing impairment and performed genetic analyses to identify causative mutations. Screening of the GJB2 gene in these cases revealed its involvement in 24 affected individuals, with specific mutations identified. For individuals without GJB2 mutations, whole exome sequencing (WES) was conducted. WES revealed 33 genetic variants, including 6 homozygous and 27 heterozygous DNA changes, two of which were previously implicated in hearing loss, while 25 variants were novel. We also observed multiple potential pathogenic heterozygous variants across different genes in some cases. Notably, a significant proportion of cases remained without potential pathogenic variants.
Conclusions: Our findings confirm the complex genetic landscape of hearing loss and the limitations of WES in achieving a 100% diagnostic rate, especially in conditions characterized by genetic heterogeneity. These results contribute to our understanding of the genetic basis of hearing loss and emphasize the need for further research and comprehensive genetic analyses to elucidate the underlying causes of this condition.
Keywords: GJB2 screening; DNA variations; Diagnostic rate; Hearing loss; Whole exome sequencing.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Azaiez H, Booth KT, Ephraim SS, Crone B, Black-Ziegelbein EA, Marini RJ, Shearer AE, Sloan-Heggen CM, Kolbe D, Casavant T, Schnieders MJ, Nishimura C, Braun T, Smith RJH. Genomic Landscape and Mutational signatures of Deafness-Associated genes. Am J Hum Genet. 2018;103(4):484–97. doi: 10.1016/j.ajhg.2018.08.006. - DOI - PMC - PubMed
-
- Bartsch O, Vatter A, Zechner U, Kohlschmidt N, Wetzig C, Baumgart A, Nospes S, Haaf T, Keilmann A. GJB2 mutations and genotype-phenotype correlation in 335 patients from Germany with nonsyndromic sensorineural hearing loss: evidence for additional recessive mutations not detected by current methods. Audiol Neurootol. 2010;15(6):375–82. doi: 10.1159/000297216. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

