Single-cell transcriptome atlases of soybean root and mature nodule reveal new regulatory programs that control the nodulation process
- PMID: 38845198
- PMCID: PMC11369782
- DOI: 10.1016/j.xplc.2024.100984
Single-cell transcriptome atlases of soybean root and mature nodule reveal new regulatory programs that control the nodulation process
Abstract
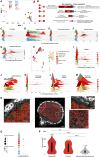
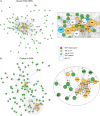
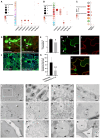
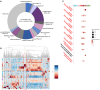
The soybean root system is complex. In addition to being composed of various cell types, the soybean root system includes the primary root, the lateral roots, and the nodule, an organ in which mutualistic symbiosis with N-fixing rhizobia occurs. A mature soybean root nodule is characterized by a central infection zone where atmospheric nitrogen is fixed and assimilated by the symbiont, resulting from the close cooperation between the plant cell and the bacteria. To date, the transcriptome of individual cells isolated from developing soybean nodules has been established, but the transcriptomic signatures of cells from the mature soybean nodule have not yet been characterized. Using single-nucleus RNA-seq and Molecular Cartography technologies, we precisely characterized the transcriptomic signature of soybean root and mature nodule cell types and revealed the co-existence of different sub-populations of B. diazoefficiens-infected cells in the mature soybean nodule, including those actively involved in nitrogen fixation and those engaged in senescence. Mining of the single-cell-resolution nodule transcriptome atlas and the associated gene co-expression network confirmed the role of known nodulation-related genes and identified new genes that control the nodulation process. For instance, we functionally characterized the role of GmFWL3, a plasma membrane microdomain-associated protein that controls rhizobial infection. Our study reveals the unique cellular complexity of the mature soybean nodule and helps redefine the concept of cell types when considering the infection zone of the soybean nodule.
Keywords: nanodomains; nodule; root; single-cell RNA-seq; soybean.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Agtuca B.J., Stopka S.A., Evans S., Samarah L., Liu Y., Xu D., Stacey M.G., Koppenaal D.W., Paša-Tolić L., Anderton C.R., et al. Metabolomic profiling of wild-type and mutant soybean root nodules using laser-ablation electrospray ionization mass spectrometry reveals altered metabolism. Plant J. 2020;103:1937–1958. - PubMed
-
- Benedito V.A., Torres-Jerez I., Murray J.D., Andriankaja A., Allen S., Kakar K., Wandrey M., Verdier J., Zuber H., Ott T., et al. A gene expression atlas of the model legume Medicago truncatula. Plant J. 2008;55:504–513. - PubMed
-
- Bergersen F. Ammonia? An Early Stable Product of Nitrogen Fixation by Soybean Root Nodules FJ Bergersen. Aust. J. Biol. Sci. 1965;18:1–9.
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

