Methods in DNA methylation array dataset analysis: A review
- PMID: 38845821
- PMCID: PMC11153885
- DOI: 10.1016/j.csbj.2024.05.015
Methods in DNA methylation array dataset analysis: A review
Abstract
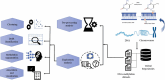
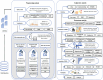
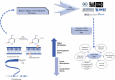
Understanding the intricate relationships between gene expression levels and epigenetic modifications in a genome is crucial to comprehending the pathogenic mechanisms of many diseases. With the advancement of DNA Methylome Profiling techniques, the emphasis on identifying Differentially Methylated Regions (DMRs/DMGs) has become crucial for biomarker discovery, offering new insights into the etiology of illnesses. This review surveys the current state of computational tools/algorithms for the analysis of microarray-based DNA methylation profiling datasets, focusing on key concepts underlying the diagnostic/prognostic CpG site extraction. It addresses methodological frameworks, algorithms, and pipelines employed by various authors, serving as a roadmap to address challenges and understand changing trends in the methodologies for analyzing array-based DNA methylation profiling datasets derived from diseased genomes. Additionally, it highlights the importance of integrating gene expression and methylation datasets for accurate biomarker identification, explores prognostic prediction models, and discusses molecular subtyping for disease classification. The review also emphasizes the contributions of machine learning, neural networks, and data mining to enhance diagnostic workflow development, thereby improving accuracy, precision, and robustness.
Keywords: Biomarker identification; Clustering; DMR analysis; Methylation segmentation; Molecular subtyping; Prognostic models.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
